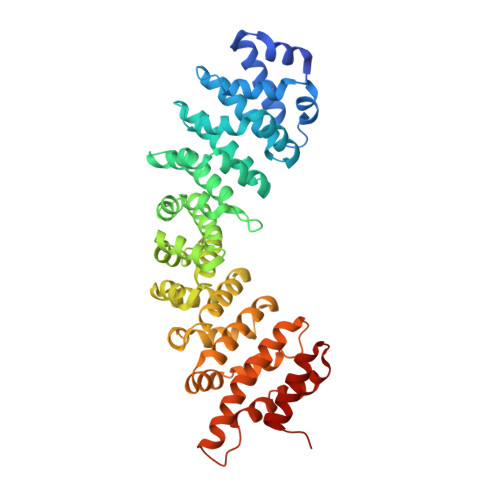
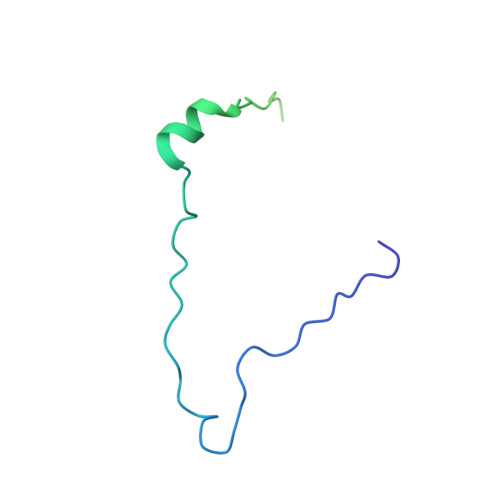
Nucleoporin Nup50 stabilizes closed conformation of armadillo repeat 10 in importin alpha 5.
Pumroy, R.A., Nardozzi, J.D., Hart, D.J., Root, M.J., Cingolani, G.(2012) J Biological Chem 287: 2022-2031
- PubMed: 22130666
- DOI: https://doi.org/10.1074/jbc.M111.315838
- Primary Citation of Related Structures:
3TJ3 - PubMed Abstract:
The human genome encodes six isoforms of importin α that show greater than 60% sequence similarity and remarkable substrate specificity. The isoform importin α5 can bind phosphorylated cargos such as STAT1 and Epstein-Barr Virus Nuclear Antigen 1, as well as the influenza virus polymerase subunit PB2. In this work, we have studied the interaction of the nucleoporin Nup50 with importin α5. We show that the first 47 residues of Nup50 bind to the C terminus of importin α5 like a "clip," stabilizing the closed conformation of ARM 10. In vitro, Nup50 binds with high affinity either to empty importin α5 or to a preassembled complex of importin α5 bound to the C-terminal domain of the import cargo PB2, resulting in a trimeric complex. By contrast, PB2 can only bind with high affinity to importin α5 in the absence of Nup50. This suggests that Nup50 primary function may not be to actively displace the import cargo from importin α5 but rather to prevent cargo rebinding in preparation for recycling. This is the first evidence for a nucleoporin modulating the import reaction by directly altering the three-dimensional structure of an import adaptor.
- Department of Biochemistry and Molecular Biology, Thomas Jefferson University, Philadelphia, Pennsylvania 19107, USA.
Organizational Affiliation: