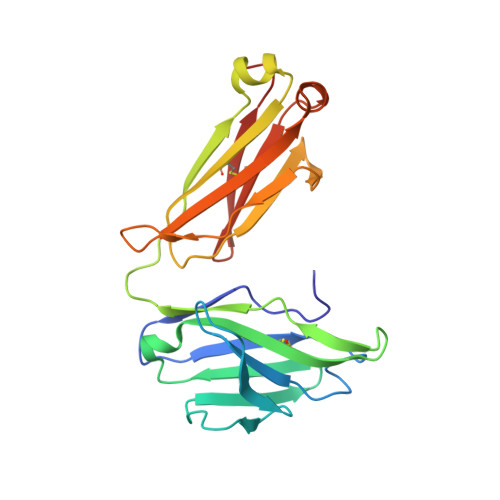
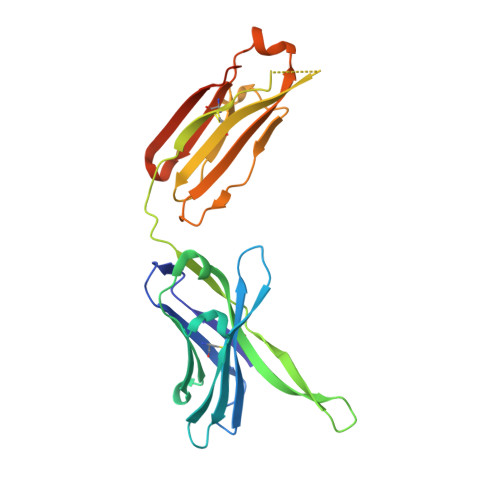
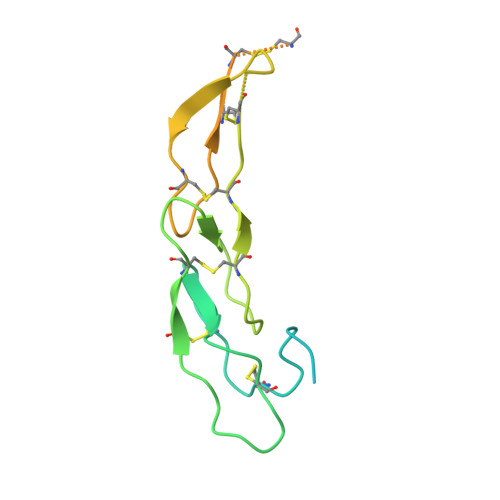
A series of Fas receptor agonist antibodies that demonstrate an inverse correlation between affinity and potency.
Chodorge, M., Zuger, S., Stirnimann, C., Briand, C., Jermutus, L., Grutter, M.G., Minter, R.R.(2012) Cell Death Differ 19: 1187-1195
- PubMed: 22261618
- DOI: https://doi.org/10.1038/cdd.2011.208
- Primary Citation of Related Structures:
3THM, 3TJE - PubMed Abstract:
Receptor agonism remains poorly understood at the molecular and mechanistic level. In this study, we identified a fully human anti-Fas antibody that could efficiently trigger apoptosis and therefore function as a potent agonist. Protein engineering and crystallography were used to mechanistically understand the agonistic activity of the antibody. The crystal structure of the complex was determined at 1.9 Å resolution and provided insights into epitope recognition and comparisons with the natural ligand FasL (Fas ligand). When we affinity-matured the agonist antibody, we observed that, surprisingly, the higher-affinity antibodies demonstrated a significant reduction, rather than an increase, in agonist activity at the Fas receptor. We propose and experimentally demonstrate a model to explain this non-intuitive impact of affinity on agonist antibody signalling and explore the implications for the discovery of therapeutic agonists in general.
- MedImmune Ltd., Granta Park, Cambridge, UK.
Organizational Affiliation: