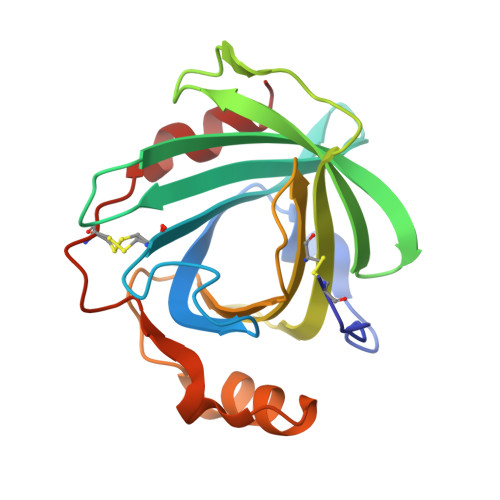
Guanidine-Ferroheme Coordination in the Mutant Protein Nitrophorin 4(L130R).
He, C., Fuchs, M.R., Ogata, H., Knipp, M.(2012) Angew Chem Int Ed Engl 51: 4470-4473
- PubMed: 22334402
- DOI: https://doi.org/10.1002/anie.201108691
- Primary Citation of Related Structures:
3TGA, 3TGB - Max-Planck-Institut für Bioanorganische Chemie, Stiftstrasse 34-36, 45470 Mülheim an der Ruhr, Germany.
Organizational Affiliation: