Structure of human SMYD2 reveals the basis of p53 tumor suppressor methylation
Wang, L., Li, L., Zhang, H., Luo, X., Dai, J., Zhou, S., Gu, J., Zhu, J., Atadja, P., Lu, C., Li, E., Zhao, K.(2011) J Biological Chem
- PubMed: 21880715
- DOI: https://doi.org/10.1074/jbc.M111.262410
- Primary Citation of Related Structures:
3TG4, 3TG5 - PubMed Abstract:
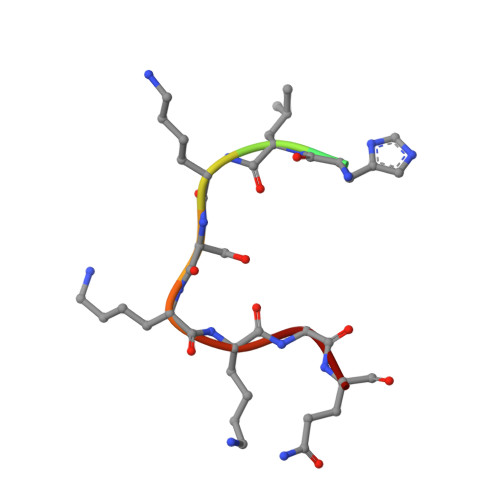
SMYD2 belongs to a subfamily of histone lysine methyltransferase and was recently identified to methylate tumor suppressor p53 and Rb. Here we report that SMYD2 prefers to methylate p53 Lys-370 over histone substrates in vitro. Consistently, the level of endogenous p53 Lys-370 monomethylation is significantly elevated when SMYD2 is overexpressed in vivo. We have solved the high resolution crystal structures of the full-length SMYD2 protein in binary complex with its cofactor S-adenosylmethionine and in ternary complex with cofactor product S-adenosylhomocysteine and p53 substrate peptide (residues 368-375), respectively. p53 peptide binds to a deep pocket of the interface between catalytic SET(1-282) and C-terminal domain (CTD) with an unprecedented U-shaped conformation. Subtle conformational change exists around the p53 binding site between the binary and ternary structures, in particular the tetratricopeptide repeat motif of the CTD. In addition, a unique EDEE motif between the loop of anti-parallel β7 and β8 sheets of the SET core not only interacts with p53 substrate but also forms a hydrogen bond network with residues from CTD. These observations suggest that the tetratricopeptide repeat and EDEE motif may play an important role in determining p53 substrate binding specificity. This is further verified by the findings that deletion of the CTD domain drastically reduces the methylation activity of SMYD2 to p53 protein. Meanwhile, mutation of EDEE residues impairs both the binding and the enzymatic activity of SMYD2 to p53 Lys-370. These data together reveal the molecular basis of SMYD2 in specifically recognizing and regulating functions of p53 tumor suppressor through Lys-370 monomethylation.
- Novartis Institutes for BioMedical Research, Building 8, Lane 898, Halei Road, Pudong, Shanghai 201203, China.
Organizational Affiliation: