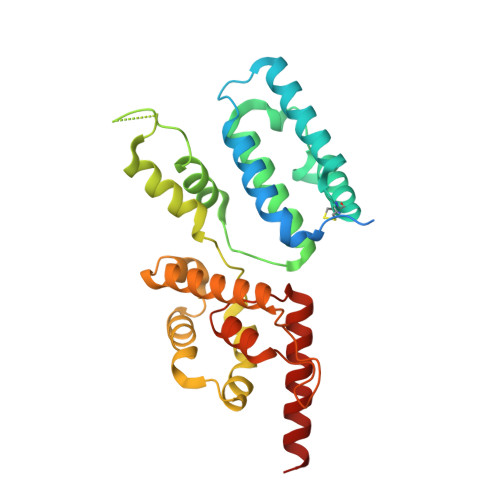
Exploring ORFan domains in giant viruses: structure of mimivirus sulfhydryl oxidase R596.
Hakim, M., Ezerina, D., Alon, A., Vonshak, O., Fass, D.(2012) PLoS One 7: e50649-e50649
- PubMed: 23209798
- DOI: https://doi.org/10.1371/journal.pone.0050649
- Primary Citation of Related Structures:
3TD7 - PubMed Abstract:
The mimivirus genome contains many genes that lack homologs in the sequence database and are thus known as ORFans. In addition, mimivirus genes that encode proteins belonging to known fold families are in some cases fused to domain-sized segments that cannot be classified. One such ORFan region is present in the mimivirus enzyme R596, a member of the Erv family of sulfhydryl oxidases. We determined the structure of a variant of full-length R596 and observed that the carboxy-terminal region of R596 assumes a folded, compact domain, demonstrating that these ORFan segments can be stable structural units. Moreover, the R596 ORFan domain fold is novel, hinting at the potential wealth of protein structural innovation yet to be discovered in large double-stranded DNA viruses. In the context of the R596 dimer, the ORFan domain contributes to formation of a broad cleft enriched with exposed aromatic groups and basic side chains, which may function in binding target proteins or localization of the enzyme within the virus factory or virions. Finally, we find evidence for an intermolecular dithiol/disulfide relay within the mimivirus R596 dimer, the first such extended, intersubunit redox-active site identified in a viral sulfhydryl oxidase.
- Department of Structural Biology, Weizmann Institute of Science, Rehovot, Israel.
Organizational Affiliation: