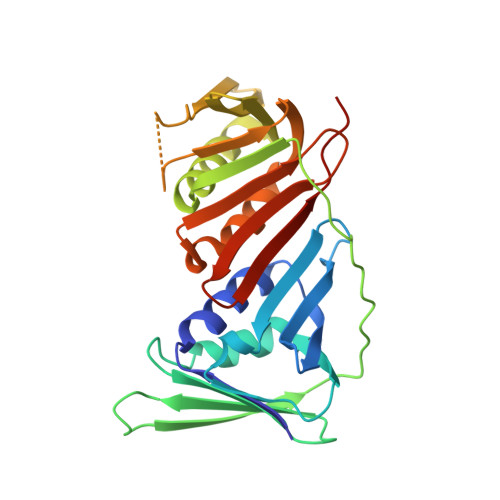
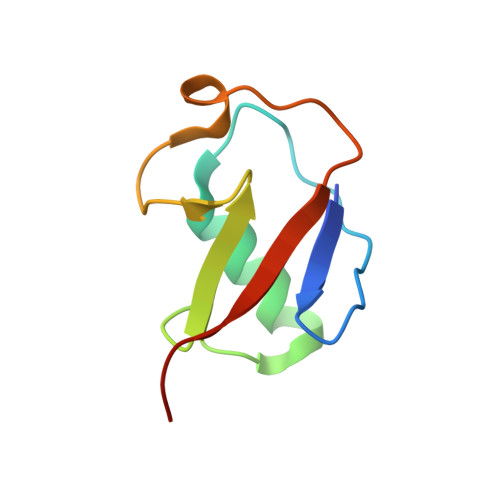
Structure of monoubiquitinated PCNA: Implications for DNA polymerase switching and Okazaki fragment maturation.
Zhang, Z., Zhang, S., Lin, S.H., Wang, X., Wu, L., Lee, E.Y., Lee, M.Y.(2012) Cell Cycle 11: 2128-2136
- PubMed: 22592530
- DOI: https://doi.org/10.4161/cc.20595
- Primary Citation of Related Structures:
3TBL - PubMed Abstract:
Ubiquitination of proliferating cell nuclear antigen (PCNA) to ub-PCNA is essential for DNA replication across bulky template lesions caused by UV radiation and alkylating agents, as ub-PCNA orchestrates the recruitment and switching of translesion synthesis (TLS) polymerases with replication polymerases. This allows replication to proceed, leaving the DNA to be repaired subsequently. Defects in a TLS polymerase, Pol η, lead to a form of Xeroderma pigmentosum, a disease characterized by severe skin sensitivity to sunlight damage and an increased incidence of skin cancer. Structurally, however, information on how ub-PCNA orchestrates the switching of these two classes of polymerases is lacking. We have solved the structure of ub-PCNA and demonstrate that the ubiquitin molecules in ub-PCNA are radially extended away from the PCNA without structural contact aside from the isopeptide bond linkage. This unique orientation provides an open platform for the recruitment of TLS polymerases through ubiquitin-interacting domains. However, the ubiquitin moieties, to the side of the equatorial PCNA plane, can place spatial constraints on the conformational flexibility of proteins bound to ub-PCNA. We show that ub-PCNA is impaired in its ability to support the coordinated actions of Fen1 and Pol δ in assays mimicking Okazaki fragment processing. This provides evidence for the novel concept that ub-PCNA may modulate additional DNA transactions other than TLS polymerase recruitment and switching.
- Department of Biochemistry and Molecular Biology, New York Medical College, Valhalla, NY, USA. Zhongtao_zhang@nymc.edu
Organizational Affiliation: