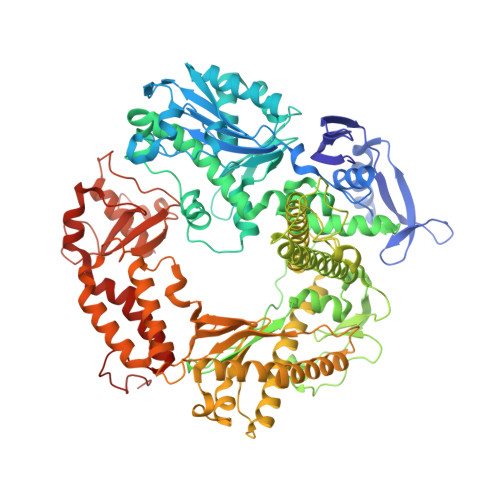
The miscoding potential of 5-hydroxycytosine arises due to template instability in the replicative polymerase active site.
Zahn, K.E., Averill, A., Wallace, S.S., Doublie, S.(2011) Biochemistry 50: 10350-10358
- PubMed: 22026756
- DOI: https://doi.org/10.1021/bi201219s
- Primary Citation of Related Structures:
3TAB, 3TAE, 3TAF, 3TAG - PubMed Abstract:
5-Hydroxycytosine (5-OHC) is a stable oxidation product of cytosine associated with an increased frequency of C → T transition mutations. When this lesion escapes recognition by the base excision repair pathway and persists to serve as a templating base during DNA synthesis, replicative DNA polymerases often misincorporate dAMP at the primer terminus, which can lead to fixation of mutations and subsequent disease. To characterize the dynamics of DNA synthesis opposite 5-OHC, we initiated a comparison of unmodified dCMP to 5-OHC, 5-fluorocytosine (5-FC), and 5-methylcytosine (5-MEC) in which these bases act as templates in the active site of RB69 gp43, a high-fidelity DNA polymerase sharing homology with human replicative DNA polymerases. This study presents the first crystal structure of any DNA polymerase binding this physiologically important premutagenic DNA lesion, showing that while dGMP is stabilized by 5-OHC through normal Watson-Crick base pairing, incorporation of dAMP leads to unstacking and instability in the template. Furthermore, the electronegativity of the C5 substituent appears to be important in the miscoding potential of these cytosine-like templates. While dAMP is incorporated opposite 5-OHC ~5 times more efficiently than opposite unmodified dCMP, an elevated level of incorporation is also observed opposite 5-FC but not 5-MEC. Taken together, these data imply that the nonuniform templating by 5-OHC is due to weakened stacking capabilities, which allows dAMP incorporation to proceed in a manner similar to that observed opposite abasic sites.
- Department of Microbiology and Molecular Genetics, University of Vermont, Burlington, Vermont 05405, United States.
Organizational Affiliation: