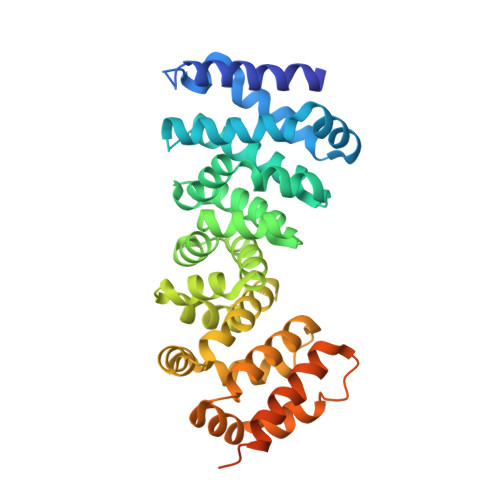
Crystal structure of the armadillo repeat domain of adenomatous polyposis coli which reveals its inherent flexibility
Zhang, Z., Lin, K., Gao, L., Chen, L., Shi, X., Wu, G.(2011) Biochem Biophys Res Commun 412: 732-736
- PubMed: 21871439
- DOI: https://doi.org/10.1016/j.bbrc.2011.08.044
- Primary Citation of Related Structures:
3T7U - PubMed Abstract:
The conserved armadillo repeat (ARM) domain of adenomatous polyposis coli (APC) protein plays an important role in the recognition of its binding partners. In this study, we report the crystal structure of APC-ARM (residues 407-775), which was determined to 2.9 Å resolution. Our structure shows that the seven armadillo repeats of APC-ARM fold together into a compact domain, with Arm2 and Arm5 presenting some deviations from canonical armadillo repeats. There is a positively charged groove on the surface of APC-ARM, which might be the recognition site for APC-binding partners. Comparison of this structure with our previously reported structure of APC (407-751), together with normal mode analysis, reveals that the APC-ARM domain possesses a limited intrinsic flexibility. We propose that this intrinsic flexibility might be an inherent property of ARM domains in general.
- State Key Laboratory of Microbial Metabolism, and School of Life Sciences & Biotechnology, Shanghai Jiao Tong University, Shanghai, China.
Organizational Affiliation: