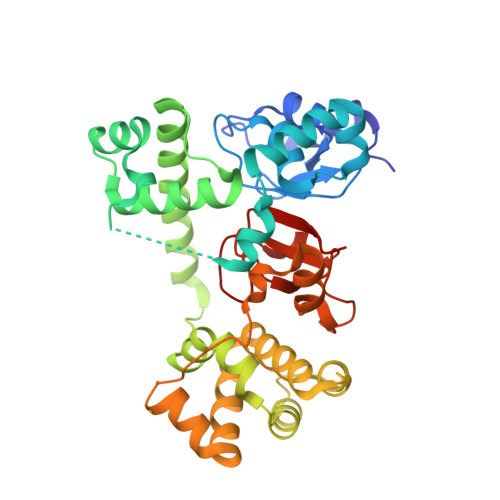
Antagonists induce a conformational change in cIAP1 that promotes autoubiquitination.
Dueber, E.C., Schoeffler, A.J., Lingel, A., Elliott, J.M., Fedorova, A.V., Giannetti, A.M., Zobel, K., Maurer, B., Varfolomeev, E., Wu, P., Wallweber, H.J., Hymowitz, S.G., Deshayes, K., Vucic, D., Fairbrother, W.J.(2011) Science 334: 376-380
- PubMed: 22021857
- DOI: https://doi.org/10.1126/science.1207862
- Primary Citation of Related Structures:
3T6P - PubMed Abstract:
Inhibitor of apoptosis (IAP) proteins are negative regulators of cell death. IAP family members contain RING domains that impart E3 ubiquitin ligase activity. Binding of endogenous or small-molecule antagonists to select baculovirus IAP repeat (BIR) domains within cellular IAP (cIAP) proteins promotes autoubiquitination and proteasomal degradation and so releases inhibition of apoptosis mediated by cIAP. Although the molecular details of antagonist-BIR domain interactions are well understood, it is not clear how this binding event influences the activity of the RING domain. Here biochemical and structural studies reveal that the unliganded, multidomain cIAP1 sequesters the RING domain within a compact, monomeric structure that prevents RING dimerization. Antagonist binding induces conformational rearrangements that enable RING dimerization and formation of the active E3 ligase.
- Department of Early Discovery Biochemistry, Genentech, 1 DNA Way, South San Francisco, CA 94080, USA.
Organizational Affiliation: