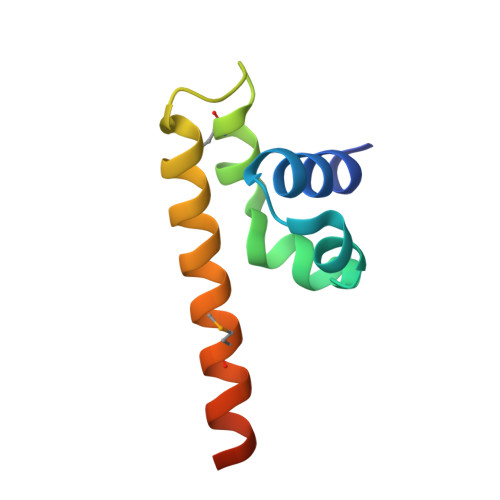
Structure of the C-terminal domain of lettuce necrotic yellows virus phosphoprotein.
Martinez, N., Ribeiro, E.A., Leyrat, C., Tarbouriech, N., Ruigrok, R.W., Jamin, M.(2013) J Virol 87: 9569-9578
- PubMed: 23785215
- DOI: https://doi.org/10.1128/JVI.00999-13
- Primary Citation of Related Structures:
3T4R - PubMed Abstract:
Lettuce necrotic yellows virus (LNYV) is a prototype of the plant-adapted cytorhabdoviruses. Through a meta-prediction of disorder, we localized a folded C-terminal domain in the amino acid sequence of its phosphoprotein. This domain consists of an autonomous folding unit that is monomeric in solution. Its structure, solved by X-ray crystallography, reveals a lollipop-shaped structure comprising five helices. The structure is different from that of the corresponding domains of other Rhabdoviridae, Filoviridae, and Paramyxovirinae; only the overall topology of the polypeptide chain seems to be conserved, suggesting that this domain evolved under weak selective pressure and varied in size by the acquisition or loss of functional modules.
- Université Grenoble Alpes, UVHCI, Grenoble, France.
Organizational Affiliation: