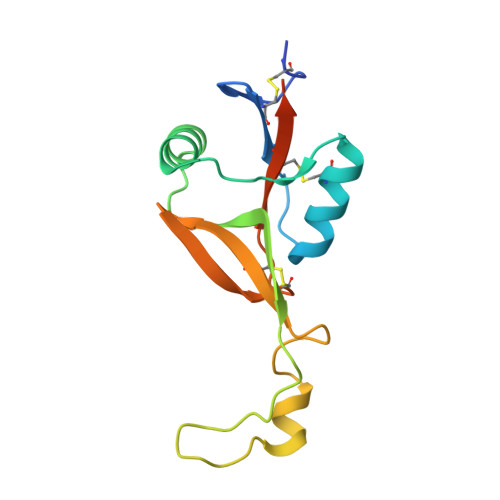
Structure of the H107R variant of the extracellular domain of mouse NKR-P1A at 2.3 A resolution.
Kolenko, P., Rozbesky, D., Vanek, O., Bezouska, K., Hasek, J., Dohnalek, J.(2011) Acta Crystallogr Sect F Struct Biol Cryst Commun 67: 1519-1523
- PubMed: 22139156
- DOI: https://doi.org/10.1107/S1744309111046203
- Primary Citation of Related Structures:
3T3A - PubMed Abstract:
The structure of the H107R variant of the extracellular domain of the mouse natural killer cell receptor NKR-P1A has been determined by X-ray diffraction at 2.3 Å resolution from a merohedrally twinned crystal. Unlike the structure of the wild-type receptor in space group I4(1)22 with a single chain per asymmetric unit, the crystals of the variant belonged to space group I4(1) with a dimer in the asymmetric unit. Different degrees of merohedral twinning were detected in five data sets collected from different crystals. The mutation does not have a significant impact on the overall structure, but led to the binding of an additional phosphate ion at the interface of the molecules.
- Institute of Macromolecular Chemistry AS CR, vvi, Prague, Czech Republic. kolenko@imc.cas.cz
Organizational Affiliation: