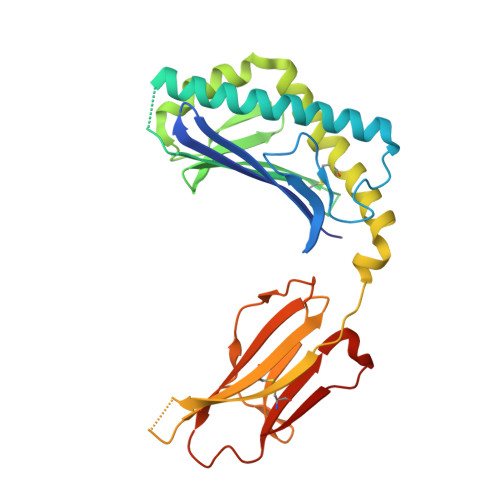
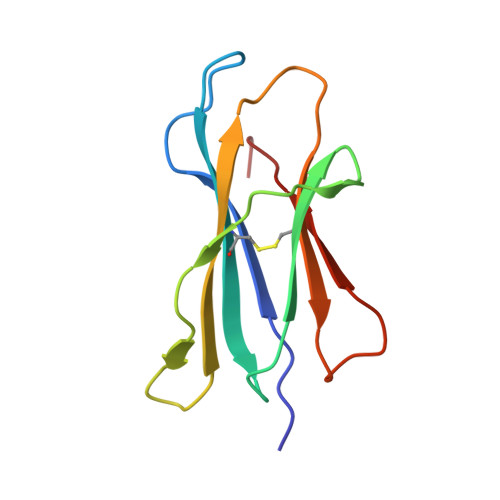
Invariant natural killer T cells recognize glycolipids from pathogenic Gram-positive bacteria.
Kinjo, Y., Illarionov, P., Vela, J.L., Pei, B., Girardi, E., Li, X., Li, Y., Imamura, M., Kaneko, Y., Okawara, A., Miyazaki, Y., Gomez-Velasco, A., Rogers, P., Dahesh, S., Uchiyama, S., Khurana, A., Kawahara, K., Yesilkaya, H., Andrew, P.W., Wong, C.H., Kawakami, K., Nizet, V., Besra, G.S., Tsuji, M., Zajonc, D.M., Kronenberg, M.(2011) Nat Immunol 12: 966-974
- PubMed: 21892173
- DOI: https://doi.org/10.1038/ni.2096
- Primary Citation of Related Structures:
3T1F - PubMed Abstract:
Natural killer T cells (NKT cells) recognize glycolipid antigens presented by CD1d. These cells express an evolutionarily conserved, invariant T cell antigen receptor (TCR), but the forces that drive TCR conservation have remained uncertain. Here we show that NKT cells recognized diacylglycerol-containing glycolipids from Streptococcus pneumoniae, the leading cause of community-acquired pneumonia, and group B Streptococcus, which causes neonatal sepsis and meningitis. Furthermore, CD1d-dependent responses by NKT cells were required for activation and host protection. The glycolipid response was dependent on vaccenic acid, which is present in low concentrations in mammalian cells. Our results show how microbial lipids position the sugar for recognition by the invariant TCR and, most notably, extend the range of microbes recognized by this conserved TCR to several clinically important bacteria.
- Division of Developmental Immunology, La Jolla Institute for Allergy & Immunology, La Jolla, California, USA.
Organizational Affiliation: