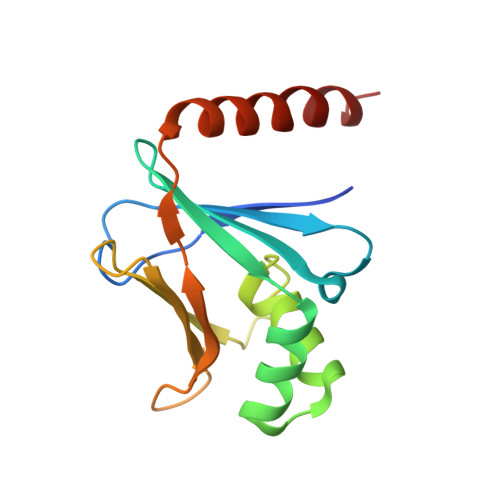
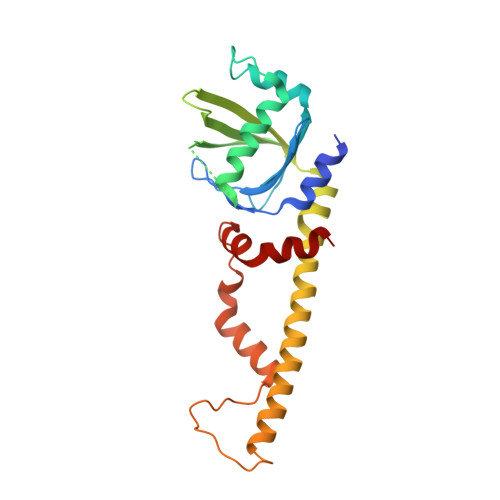
XRCC4 Protein Interactions with XRCC4-like Factor (XLF) Create an Extended Grooved Scaffold for DNA Ligation and Double Strand Break Repair.
Hammel, M., Rey, M., Yu, Y., Mani, R.S., Classen, S., Liu, M., Pique, M.E., Fang, S., Mahaney, B.L., Weinfeld, M., Schriemer, D.C., Lees-Miller, S.P., Tainer, J.A.(2011) J Biological Chem 286: 32638-32650
- PubMed: 21775435
- DOI: https://doi.org/10.1074/jbc.M111.272641
- Primary Citation of Related Structures:
3SR2 - PubMed Abstract:
The XRCC4-like factor (XLF)-XRCC4 complex is essential for nonhomologous end joining, the major repair pathway for DNA double strand breaks in human cells. Yet, how XLF binds XRCC4 and impacts nonhomologous end joining functions has been enigmatic. Here, we report the XLF-XRCC4 complex crystal structure in combination with biophysical and mutational analyses to define the XLF-XRCC4 interactions. Crystal and solution structures plus mutations characterize alternating XRCC4 and XLF head domain interfaces forming parallel super-helical filaments. XLF Leu-115 ("Leu-lock") inserts into a hydrophobic pocket formed by XRCC4 Met-59, Met-61, Lys-65, Lys-99, Phe-106, and Leu-108 in synergy with pseudo-symmetric β-zipper hydrogen bonds to drive specificity. XLF C terminus and DNA enhance parallel filament formation. Super-helical XLF-XRCC4 filaments form a positively charged channel to bind DNA and align ends for efficient ligation. Collective results reveal how human XLF and XRCC4 interact to bind DNA, suggest consequences of patient mutations, and support a unified molecular mechanism for XLF-XRCC4 stimulation of DNA ligation.
- Physical Biosciences Division, Lawrence Berkeley National Laboratory, Berkeley, California 94720, USA.
Organizational Affiliation: