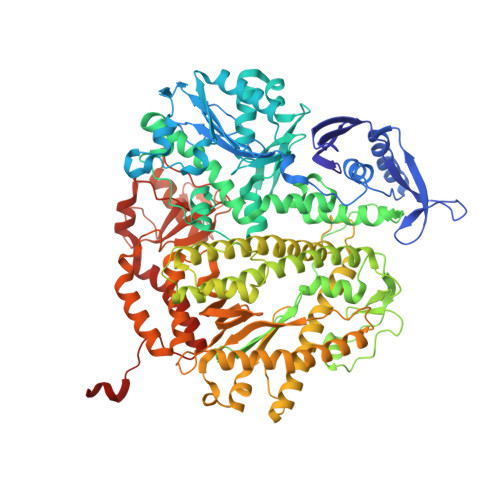
Structural Insights into Complete Metal Ion Coordination from Ternary Complexes of B Family RB69 DNA Polymerase.
Xia, S., Wang, M., Blaha, G., Konigsberg, W.H., Wang, J.(2011) Biochemistry 50: 9114-9124
- PubMed: 21923197
- DOI: https://doi.org/10.1021/bi201260h
- Primary Citation of Related Structures:
3S9H, 3SCX, 3SI6, 3SJJ, 3SNN, 3SPY, 3SPZ, 3SQ0, 3SQ1 - PubMed Abstract:
We have captured a preinsertion ternary complex of RB69 DNA polymerase (RB69pol) containing the 3' hydroxyl group at the terminus of an extendable primer (ptO3') and a nonhydrolyzable 2'-deoxyuridine 5'-α,β-substituted triphosphate, dUpXpp, where X is either NH or CH(2), opposite a complementary templating dA nucleotide residue. Here we report four structures of these complexes formed by three different RB69pol variants with catalytically inert Ca(2+) and four other structures with catalytically competent Mn(2+) or Mg(2+). These structures provide new insights into why the complete divalent metal-ion coordination complexes at the A and B sites are required for nucleotidyl transfer. They show that the metal ion in the A site brings ptO3' close to the α-phosphorus atom (Pα) of the incoming dNTP to enable phosphodiester bond formation through simultaneous coordination of both ptO3' and the nonbridging Sp oxygen of the dNTP's α-phosphate. The coordination bond length of metal ion A as well as its ionic radius determines how close ptO3' can approach Pα. These variables are expected to affect the rate of bond formation. The metal ion in the B site brings the pyrophosphate product close enough to Pα to enable pyrophosphorolysis and assist in the departure of the pyrophosphate. In these dUpXpp-containing complexes, ptO3' occupies the vertex of a distorted metal ion A coordination octahedron. When ptO3' is placed at the vertex of an undistorted, idealized metal ion A octahedron, it is within bond formation distance to Pα. This geometric relationship appears to be conserved among DNA polymerases of known structure.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, Connecticut 96520-8114, United States.
Organizational Affiliation: