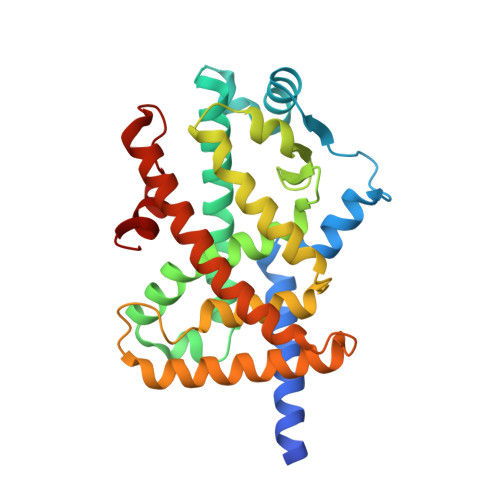
Structural basis for iloprost as a dual peroxisome proliferator-activated receptor alpha/delta agonist.
Jin, L., Lin, S., Rong, H., Zheng, S., Jin, S., Wang, R., Li, Y.(2011) J Biological Chem 286: 31473-31479
- PubMed: 21775429
- DOI: https://doi.org/10.1074/jbc.M111.266023
- Primary Citation of Related Structures:
3SP6, 3SP9 - PubMed Abstract:
Iloprost is a prostacyclin analog that has been used to treat many vascular conditions. Peroxisome proliferator-activated receptors (PPARs) are ligand-regulated transcription factors with various important biological effects such as metabolic and cardiovascular physiology. Here, we report the crystal structures of the PPARα ligand-binding domain and PPARδ ligand-binding domain bound to iloprost, thus providing unambiguous evidence for the direct interaction between iloprost and PPARs and a structural basis for the recognition of PPARα/δ by this prostacyclin analog. In addition to conserved contacts for all PPARα ligands, iloprost also initiates several specific interactions with PPARs using its unique structural groups. Structural and functional studies of receptor-ligand interactions reveal strong functional correlations of the iloprost-PPARα/δ interactions as well as the molecular basis of PPAR subtype selectivity toward iloprost ligand. As such, the structural mechanism may provide a more rational template for designing novel compounds targeting PPARs with more favorable pharmacologic impact based on existing iloprost drugs.
- State Key Laboratory of Cellular Stress Biology, School of Life Sciences, Xiamen University, Fujian 361005, China.
Organizational Affiliation: