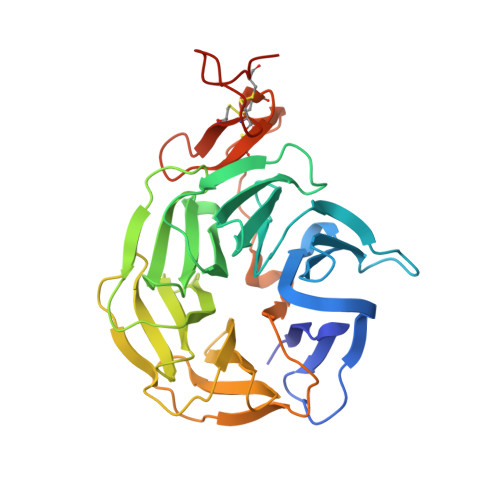
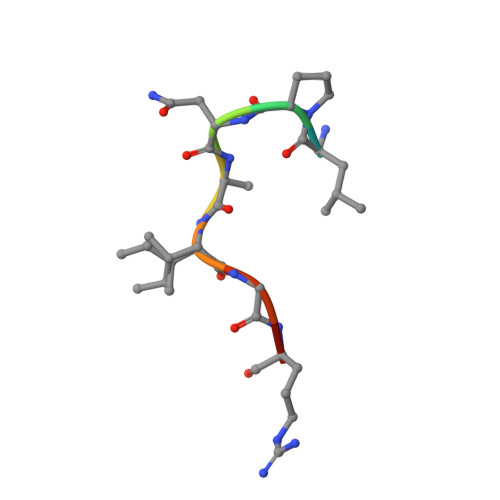
Wnt antagonists bind through a short peptide to the first beta-propeller domain of LRP5/6.
Bourhis, E., Wang, W., Tam, C., Hwang, J., Zhang, Y., Spittler, D., Huang, O.W., Gong, Y., Estevez, A., Zilberleyb, I., Rouge, L., Chiu, C., Wu, Y., Costa, M., Hannoush, R.N., Franke, Y., Cochran, A.G.(2011) Structure 19: 1433-1442
- PubMed: 21944579
- DOI: https://doi.org/10.1016/j.str.2011.07.005
- Primary Citation of Related Structures:
3SOB, 3SOQ, 3SOV - PubMed Abstract:
The Wnt pathway inhibitors DKK1 and sclerostin (SOST) are important therapeutic targets in diseases involving bone loss or damage. It has been appreciated that Wnt coreceptors LRP5/6 are also important, as human missense mutations that result in bone overgrowth (bone mineral density, or BMD, mutations) cluster to the E1 propeller domain of LRP5. Here, we report a crystal structure of LRP6 E1 bound to an antibody, revealing that the E1 domain is a peptide recognition module. Remarkably, the consensus E1 binding sequence is a close match to a conserved tripeptide motif present in all Wnt inhibitors that bind LRP5/6. We show that this motif is important for DKK1 and SOST binding to LRP6 and for inhibitory function, providing a detailed structural explanation for the effect of the BMD mutations.
- Department of Early Discovery Biochemistry, Genentech Research and Early Development, 1 DNA Way, South San Francisco, CA 94080, USA.
Organizational Affiliation: