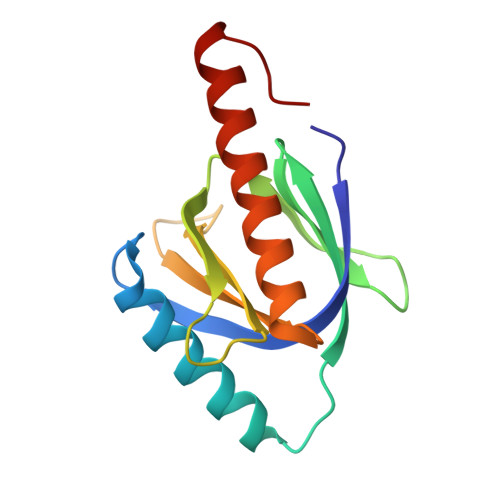
Atomic structure of the autosomal recessive hypercholesterolemia phosphotyrosine-binding domain in complex with the LDL-receptor tail.
Dvir, H., Shah, M., Girardi, E., Guo, L., Farquhar, M.G., Zajonc, D.M.(2012) Proc Natl Acad Sci U S A 109: 6916-6921
- PubMed: 22509010
- DOI: https://doi.org/10.1073/pnas.1114128109
- Primary Citation of Related Structures:
3SO6 - PubMed Abstract:
Hypercholesterolemia, high serum cholesterol in the form of LDL, is a major risk factor for atherosclerosis. LDL is mostly degraded in the liver after its cellular internalization with the LDL receptor (LDLR). This clathrin-mediated endocytosis depends on the protein autosomal recessive hypercholesterolemia (ARH), which binds the LDLR cytoplasmic tail. Mutations in either the LDLR tail or in ARH lead to hypercholesterolemia and premature atherosclerosis. Despite the significance of this interaction for cholesterol homeostasis, no structure of either ARH or the LDLR tail is available to determine its molecular basis. We report the crystal structure at 1.37-Å resolution of the phosphotyrosine-binding (PTB) domain of ARH in complex with an LDLR tail peptide containing the FxNPxY(0) internalization signal. Surprisingly, ARH interacts with a longer portion of the tail than previously recognized, which extends to I(-7)xF(-5)xNPxY(0)QK(+2). The LDLR tail assumes a unique "Hook"-like structure with a double β-turn conformation, which is accommodated in distinctive ARH structural determinants (i.e., an extended backbone hydrogen-bonding platform, three hydrophobic helical grooves, and a hydrophobic pocket for Y(0)). This unique complementarity differs significantly in related PTB proteins and may account for the unique physiological role of these partners in the hepatic uptake of cholesterol LDL. Moreover, the unusual hydrophobic pocket for Y(0) explains the distinctive ability of ARH to internalize proteins containing either FxNPxY(0) or FxNPxF(0) sequences. Biophysical measurements reveal how mutations associated with hypercholesterolemia destabilize ARH and its complex with LDLR and illuminate LDL internalization defects seen in patients.
- Division of Cell Biology, La Jolla Institute for Allergy and Immunology, La Jolla, CA 92037, USA.
Organizational Affiliation: