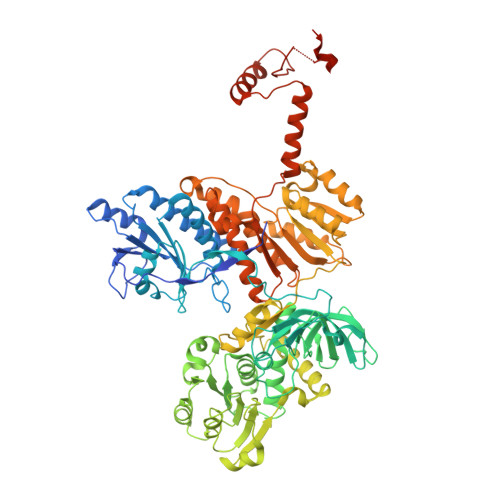
Divergence of multimodular polyketide synthases revealed by a didomain structure.
Zheng, J., Gay, D.C., Demeler, B., White, M.A., Keatinge-Clay, A.T.(2012) Nat Chem Biol 8: 615-621
- PubMed: 22634636
- DOI: https://doi.org/10.1038/nchembio.964
- Primary Citation of Related Structures:
3SLK - PubMed Abstract:
The enoylreductase (ER) is the final common enzyme from modular polyketide synthases (PKSs) to be structurally characterized. The 3.0 Å-resolution structure of the didomain comprising the ketoreductase (KR) and ER from the second module of the spinosyn PKS reveals that ER shares an ∼600-Å(2) interface with KR distinct from that of the related mammalian fatty acid synthase (FAS). In contrast to the ER domains of the mammalian FAS, the ER domains of the second module of the spinosyn PKS do not make contact across the two-fold axis of the synthase. This monomeric organization may have been necessary in the evolution of multimodular PKSs to enable acyl carrier proteins to access each of their cognate enzymes. The isolated ER domain showed activity toward a substrate analog, enabling us to determine the contributions of its active site residues.
- Department of Chemistry and Biochemistry, The University of Texas at Austin, Austin, Texas, USA.
Organizational Affiliation: