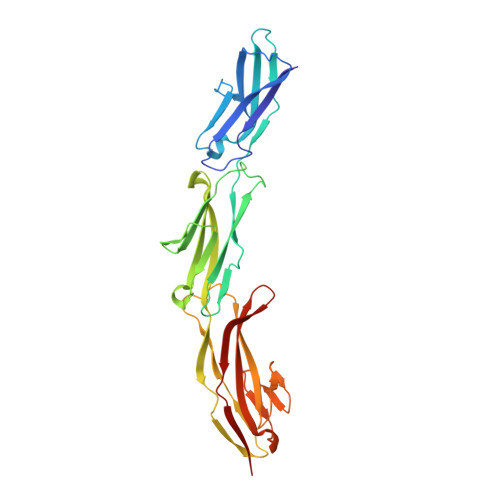
Structure of the three N-terminal immunoglobulin domains of the highly immunogenic outer capsid protein from a T4-like bacteriophage.
Fokine, A., Islam, M.Z., Zhang, Z., Bowman, V.D., Rao, V.B., Rossmann, M.G.(2011) J Virol 85: 8141-8148
- PubMed: 21632759
- DOI: https://doi.org/10.1128/JVI.00847-11
- Primary Citation of Related Structures:
3SHS - PubMed Abstract:
The head of bacteriophage T4 is decorated with 155 copies of the highly antigenic outer capsid protein (Hoc). One Hoc molecule binds near the center of each hexameric capsomer. Hoc is dispensable for capsid assembly and has been used to display pathogenic antigens on the surface of T4. Here we report the crystal structure of a protein containing the first three of four domains of Hoc from bacteriophage RB49, a close relative of T4. The structure shows an approximately linear arrangement of the protein domains. Each of these domains has an immunoglobulin-like fold, frequently found in cell attachment molecules. In addition, we report biochemical data suggesting that Hoc can bind to Escherichia coli, supporting the hypothesis that Hoc could attach the phage capsids to bacterial surfaces and perhaps also to other organisms. The capacity for such reversible adhesion probably provides survival advantages to the bacteriophage.
- Department of Biological Sciences, Purdue University, West Lafayette, Indiana 47907-2032, USA.
Organizational Affiliation: