The development of Peptide-based tools for the analysis of angiogenesis.
Fedorova, A., Zobel, K., Gill, H.S., Ogasawara, A., Flores, J.E., Tinianow, J.N., Vanderbilt, A.N., Wu, P., Meng, Y.G., Williams, S.P., Wiesmann, C., Murray, J., Marik, J., Deshayes, K.(2011) Chem Biol 18: 839-845
- PubMed: 21802005
- DOI: https://doi.org/10.1016/j.chembiol.2011.05.011
- Primary Citation of Related Structures:
3S1B, 3S1K - PubMed Abstract:
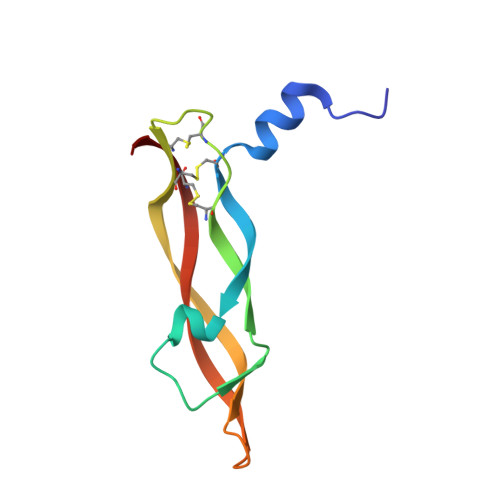
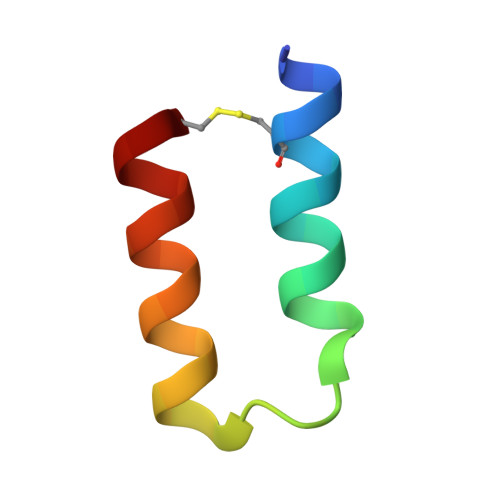
Limitations to the application of molecularly targeted cancer therapies are the inability to accurately match patient with effective treatment and the absence of a prompt readout of posttreatment response. Noninvasive agents that rapidly report vascular endothelial growth factor (VEGF) levels using positron emission tomography (PET) have the potential to enhance anti-angiogenesis therapies. Using phage display, two distinct classes of peptides were identified that bind to VEGF with nanomolar affinity and high selectivity. Co-crystal structures of these different peptide classes demonstrate that both bind to the receptor-binding region of VEGF. (18)F-radiolabelling of these peptides facilitated the acquisition of PET images of tumor VEGF levels in a HM7 xenograph model. The images obtained from one 59-residue probe, (18)F-Z-3B, 2 hr postinjection are comparable to those obtained with anti-VEGF antibody B20 72 hr postinjection. Furthermore, VEGF levels in growing SKOV3 tumors were followed using (18)F-Z-3B as a PET probe with VEGF levels increasing with tumor size.
- Department of Early Discovery Biochemistry, Genentech, South San Francisco, CA 94080, USA.
Organizational Affiliation: