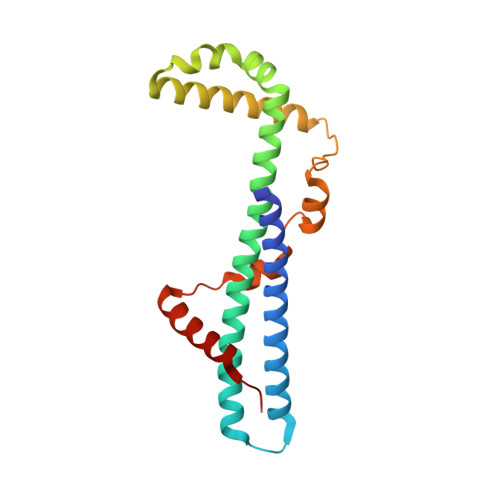
The first structure of polarity suppression protein, Psu from enterobacteria phage P4, reveals a novel fold and a knotted dimer
Banerjee, R., Nath, S., Ranjan, A., Khamrui, S., Pani, B., Sen, R., Sen, U.(2012) J Biological Chem 287: 44667-44675
- PubMed: 23150672
- DOI: https://doi.org/10.1074/jbc.M112.423202
- Primary Citation of Related Structures:
3RX6, 4DVD - PubMed Abstract:
Psu is a capsid decoration protein of bacteriophage P4 and acts as an antiterminator of Rho-dependent transcription termination in bacteria. So far, no structures have been reported for the Psu protein or its homologues. Here, we report the first structure of Psu solved by the Hg(2+) single wavelength anomalous dispersion method, which reveals that Psu exists as a knotted homodimer and is first of its kind in nature. Each monomer of Psu attains a novel fold around a tight coiled-coil motif. CD spectroscopy and the structure of an engineered disulfide-bridged Psu derivative reveal that the protein folds reversibly and reassembles by itself into the knotted dimeric conformation without the requirement of any chaperone. This structure would help to explain the functional properties of the protein and can be used as a template to design a minimal peptide fragment that can be used as a drug against Rho-dependent transcription termination in bacteria.
- Crystallography and Molecular Biology Division, Saha Institute of Nuclear Physics, 1/AF, Bidhannagar, Kolkata 700064, India.
Organizational Affiliation: