Insights into noncanonical E1 enzyme activation from the structure of autophagic E1 Atg7 with Atg8.
Hong, S.B., Kim, B.W., Lee, K.E., Kim, S.W., Jeon, H., Kim, J., Song, H.K.(2011) Nat Struct Mol Biol 18: 1323-1330
- PubMed: 22056771
- DOI: https://doi.org/10.1038/nsmb.2165
- Primary Citation of Related Structures:
3RUI, 3RUJ - PubMed Abstract:
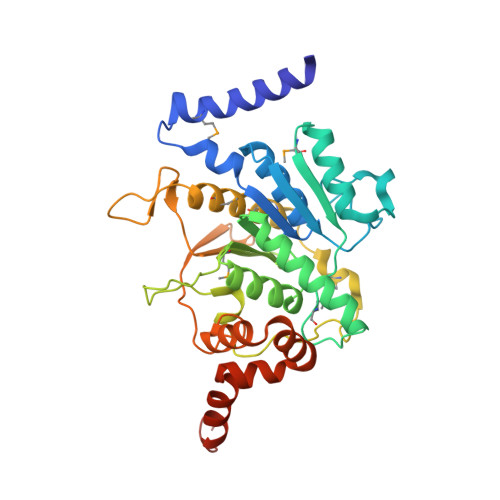
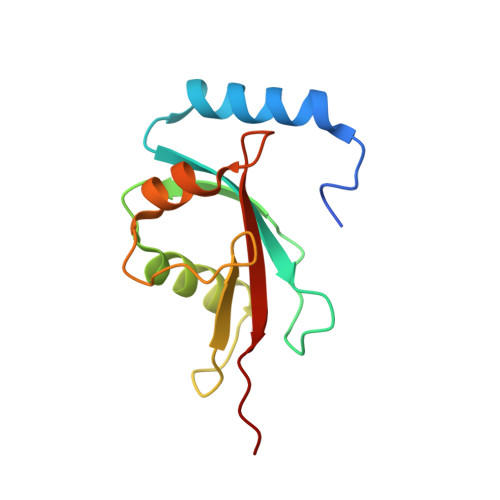
Autophagy is the degradation of cellular organelles via the lysosomal pathway. The autophagic ubiquitin-like (Ubl) molecule Atg8 is activated by the E1-like enzyme Atg7. As this noncanonical E1 enzyme's domain organization is unique among Ubl-activating E1 enzymes, the structural basis for its interactions with Atg8 and partner E2 enzymes remains obscure. Here we present the structure of the N-terminal domain of Atg7, revealing a unique protein fold and interactions with both autophagic E2 enzymes Atg3 and Atg10. The structure of the C-terminal domain of Atg7 in complex with Atg8 shows the mode of dimerization and mechanism of recognition of Atg8. Notably, the catalytic cysteine residue in Atg7 is positioned close to the C-terminal glycine of Atg8, its target for thioester formation, potentially eliminating the need for large conformational rearrangements characteristic of other E1s.
- School of Life Sciences and Biotechnology, Korea University, Seoul, South Korea.
Organizational Affiliation: