Crystal structure of the Bowman-Birk serine protease inhibitor BTCI in complex with trypsin and chymotrypsin
Esteves, G.F., Teles, R.C.L., Cavalcante, N.S., Neves, D., Ventura, M.M., Barbosa, J.A.R.G., Freitas, S.M.To be published.
Experimental Data Snapshot
Starting Models: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cationic trypsin | A [auth T] | 223 | Bos taurus | Mutation(s): 0 EC: 3.4.21.4 | 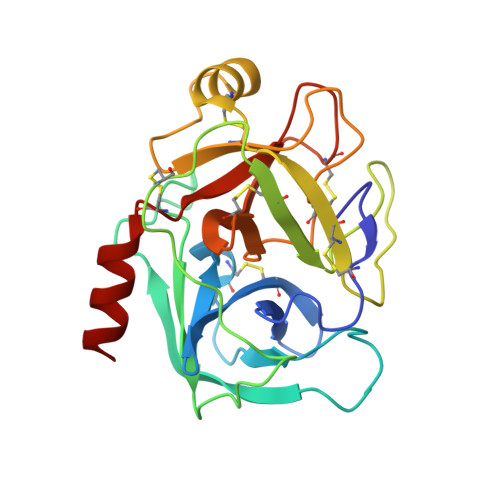 |
UniProt | |||||
Find proteins for P00760 (Bos taurus) Explore P00760 Go to UniProtKB: P00760 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00760 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Bowman-Birk type seed trypsin and chymotrypsin inhibitor | 61 | Vigna unguiculata | Mutation(s): 0 | 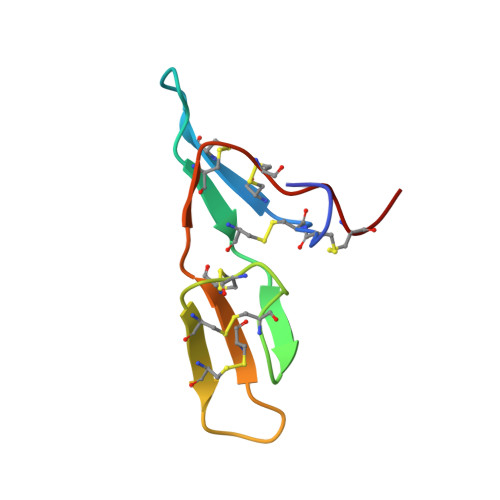 | |
UniProt | |||||
Find proteins for P17734 (Vigna unguiculata) Explore P17734 Go to UniProtKB: P17734 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P17734 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Chymotrypsinogen A | 11 | Bos taurus | Mutation(s): 0 EC: 3.4.21.1 |  | |
UniProt | |||||
Find proteins for P00766 (Bos taurus) Explore P00766 Go to UniProtKB: P00766 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00766 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Chymotrypsinogen A | 131 | Bos taurus | Mutation(s): 0 EC: 3.4.21.1 | 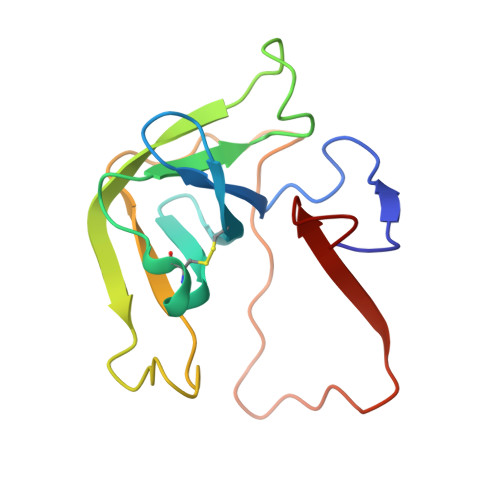 | |
UniProt | |||||
Find proteins for P00766 (Bos taurus) Explore P00766 Go to UniProtKB: P00766 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00766 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Chymotrypsinogen A | 96 | Bos taurus | Mutation(s): 0 EC: 3.4.21.1 | 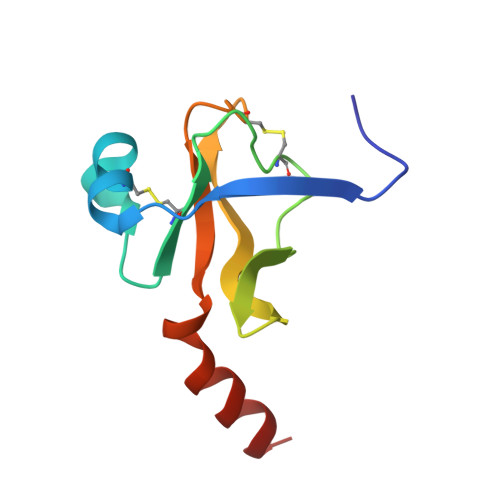 | |
UniProt | |||||
Find proteins for P00766 (Bos taurus) Explore P00766 Go to UniProtKB: P00766 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00766 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MRD Query on MRD | R [auth T], Y [auth D] | (4R)-2-METHYLPENTANE-2,4-DIOL C6 H14 O2 SVTBMSDMJJWYQN-RXMQYKEDSA-N |  | ||
| SO4 Query on SO4 | AA [auth E] L [auth T] M [auth T] N [auth T] T [auth B] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| GOL Query on GOL | G [auth T] H [auth T] I [auth T] J [auth T] K [auth T] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | BA [auth E] CA [auth E] O [auth T] P [auth T] Q [auth T] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| CA Query on CA | F [auth T] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 49.477 | α = 67.28 |
| b = 54.571 | β = 71.04 |
| c = 69.286 | γ = 73.55 |
| Software Name | Purpose |
|---|---|
| MOLREP | phasing |
| REFMAC | refinement |
| HKL-2000 | data reduction |
| SCALEPACK | data scaling |