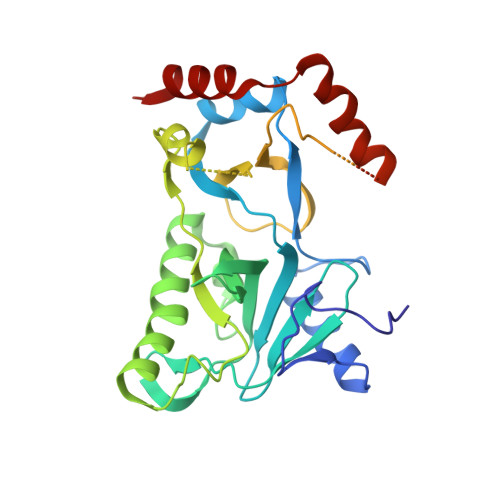
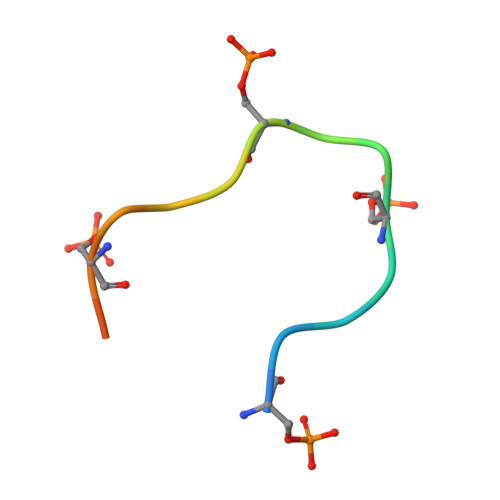
Structural insights to how mammalian capping enzyme reads the CTD code.
Ghosh, A., Shuman, S., Lima, C.D.(2011) Mol Cell 43: 299-310
- PubMed: 21683636
- DOI: https://doi.org/10.1016/j.molcel.2011.06.001
- Primary Citation of Related Structures:
3RTX - PubMed Abstract:
Physical interaction between the phosphorylated RNA polymerase II carboxyl-terminal domain (CTD) and cellular capping enzymes is required for efficient formation of the 5' mRNA cap, the first modification of nascent mRNA. Here, we report the crystal structure of the RNA guanylyltransferase component of mammalian capping enzyme (Mce) bound to a CTD phosphopeptide. The CTD adopts an extended β-like conformation that docks Tyr1 and Ser5-PO(4) onto the Mce nucleotidyltransferase domain. Structure-guided mutational analysis verified that the Mce-CTD interface is a tunable determinant of CTD binding and stimulation of guanylyltransferase activity, and of Mce function in vivo. The location and composition of the CTD binding site on mammalian capping enzyme is distinct from that of a yeast capping enzyme that recognizes the same CTD primary structure. Thus, capping enzymes from different taxa have evolved different strategies to read the CTD code.
- Structural Biology Program, Sloan-Kettering Institute, New York, NY 10065, USA.
Organizational Affiliation: