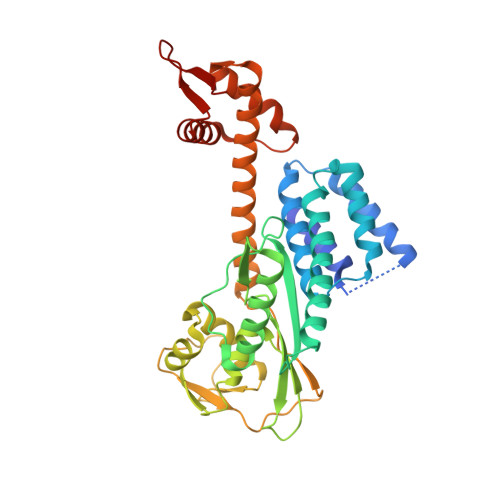
A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases.
Calabrese, M.F., Scott, D.C., Duda, D.M., Grace, C.R., Kurinov, I., Kriwacki, R.W., Schulman, B.A.(2011) Nat Struct Mol Biol 18: 947-949
- PubMed: 21765416
- DOI: https://doi.org/10.1038/nsmb.2086
- Primary Citation of Related Structures:
3RTR - PubMed Abstract:
How RING E3 ligases mediate E2-to-substrate ubiquitin-like protein (UBL) transfer remains unknown. Here we address how the RING E3 RBX1 positions NEDD8's E2 (UBC12) and substrate (CUL1). We find that existing structures are incompatible with CUL1 NEDD8ylation and report a new conformation of RBX1 that places UBC12 adjacent to CUL1. We propose RING domain rotation as a general mechanism for UBL transfer for the largest family of E3s.
- Department of Structural Biology, St. Jude Children's Research Hospital, Memphis, Tennessee, USA. matthew.calabrese@stjude.org
Organizational Affiliation: