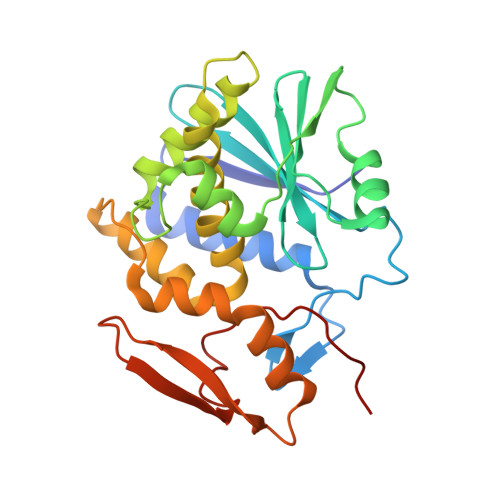
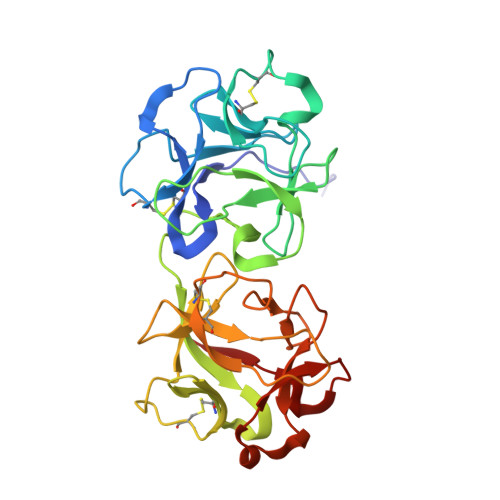
X-ray analysis of substrate analogs in the ricin A-chain active site.
Monzingo, A.F., Robertus, J.D.(1992) J Mol Biology 227: 1136-1145
- PubMed: 1433290
- DOI: https://doi.org/10.1016/0022-2836(92)90526-p
- Primary Citation of Related Structures:
3RTI, 3RTJ - PubMed Abstract:
Ricin A-chain is an N-glycosidase that hydrolyzes the adenine ring from a specific adenosine of rRNA. Formycin monophosphate (FMP) and adenyl(3'-->5')guanosine (ApG) were bound to ricin A-chain and their structures elucidated by X-ray crystallography. The formycin ring stacks between tyrosines 80 and 123 and at least four hydrogen bonds are made to the adenine moiety. A residue invariant in this enzyme class, Arg180, appears to hydrogen bond to N-3 of the susceptible adenine. Three hypothetical models for binding a true hexanucleotide substrate, CGAGAG, are proposed. They incorporate adenine binding, shown by crystallography, but also include geometry likely to favor catalysis. For example, efforts have been made to orient the ribose ring in a way that allows solvent attack and oxycarbonium stabilization by the enzyme. The favored model is a simple perturbation of the tetraloop structure determined by nuclear magnetic resonance for similar polynucleotides. The model is attractive in that specific roles are defined for conserved protein residues. A mechanism of action is proposed. It invokes oxycarbonium ion stabilization on ribose by Glu177 in the transition state. Arg180 stabilizes anion development on the leaving adenine by protonation at N-3 and may activate a trapped water molecule that is the ultimate nucleophile in the depurination.
- Clayton Foundation Biochemical Institute, Department of Chemistry and Biochemistry, University of Texas, Austin 78712.
Organizational Affiliation: