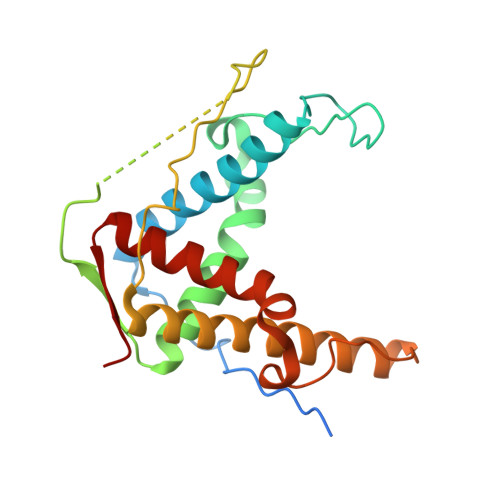
Structural determination of the phosphorylation domain of the ryanodine receptor.
Sharma, P., Ishiyama, N., Nair, U., Li, W., Dong, A., Miyake, T., Wilson, A., Ryan, T., Maclennan, D.H., Kislinger, T., Ikura, M., Dhe-Paganon, S., Gramolini, A.O.(2012) FEBS J 279: 3952-3964
- PubMed: 22913516
- DOI: https://doi.org/10.1111/j.1742-4658.2012.08755.x
- Primary Citation of Related Structures:
3RQR - PubMed Abstract:
The ryanodine receptor (RyR) is a large, homotetrameric sarcoplasmic reticulum membrane protein that is essential for Ca(2+) cycling in both skeletal and cardiac muscle. Genetic mutations in RyR1 are associated with severe conditions including malignant hyperthermia (MH) and central core disease. One phosphorylation site (Ser 2843) has been identified in a segment of RyR1 flanked by two RyR motifs, which are found exclusively in all RyR isoforms as closely associated tandem (or paired) motifs, and are named after the protein itself. These motifs also contain six known MH mutations. In this study, we designed, expressed and purified the tandem RyR motifs, and show that this domain contains a putative binding site for the Ca(2+)/calmodulin-dependent protein kinase β isoform. We present a 2.2 Å resolution crystal structure of the RyR domain revealing a two-fold, symmetric, extended four-helix bundle stabilized by a β sheet. Using mathematical modelling, we fit our crystal structure within a tetrameric electron microscopy (EM) structure of native RyR1, and propose that this domain is localized in the RyR clamp region, which is absent in its cousin protein inositol 1,4,5-trisphosphate receptor.
- Department of Physiology, University of Toronto, Toronto, Ontario, Canada.
Organizational Affiliation: