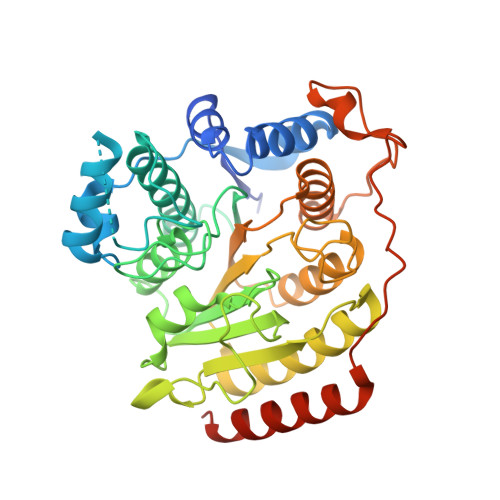
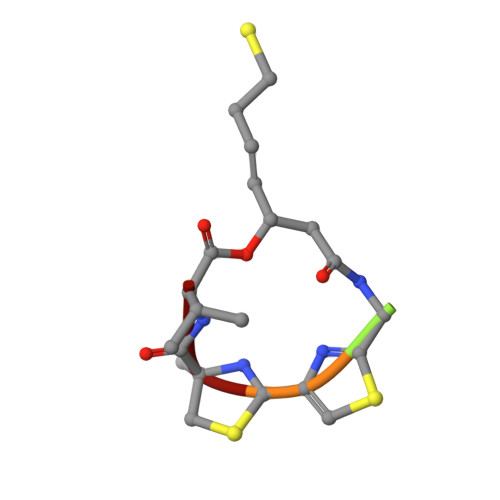
Structural basis of the antiproliferative activity of largazole, a depsipeptide inhibitor of the histone deacetylases.
Cole, K.E., Dowling, D.P., Boone, M.A., Phillips, A.J., Christianson, D.W.(2011) J Am Chem Soc 133: 12474-12477
- PubMed: 21790156
- DOI: https://doi.org/10.1021/ja205972n
- Primary Citation of Related Structures:
3RQD - PubMed Abstract:
Largazole is a macrocyclic depsipeptide originally isolated from the marine cyanobacterium Symploca sp., which is indigenous to the warm, blue-green waters of Key Largo, Florida (whence largazole derives its name). Largazole contains an unusual thiazoline-thiazole ring system that rigidifies its macrocyclic skeleton, and it also contains a lipophilic thioester side chain. Hydrolysis of the thioester in vivo yields largazole thiol, which exhibits remarkable antiproliferative effects and is believed to be the most potent inhibitor of the metal-dependent histone deacetylases (HDACs). Here, the 2.14 Å-resolution crystal structure of the HDAC8-largazole thiol complex is the first of an HDAC complexed with a macrocyclic inhibitor and reveals that ideal thiolate-zinc coordination geometry is the key chemical feature responsible for its exceptional affinity and biological activity. Notably, the core structure of largazole is conserved in romidepsin, a depsipeptide natural product formulated as the drug Istodax recently approved for cancer chemotherapy. Accordingly, the structure of the HDAC8-largazole thiol complex is the first to illustrate the mode of action of a new class of therapeutically important HDAC inhibitors.
- Roy and Diana Vagelos Laboratories, Department of Chemistry, University of Pennsylvania, 231 South 34th Street, Philadelphia, Pennsylvania 19104-6323, United States.
Organizational Affiliation: