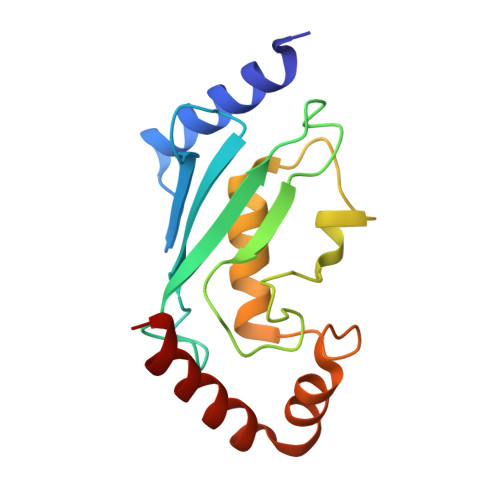
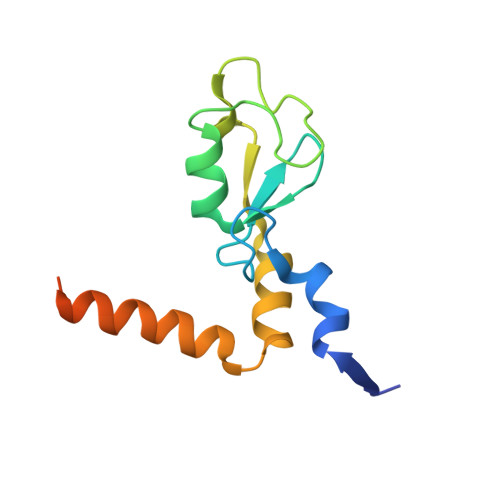
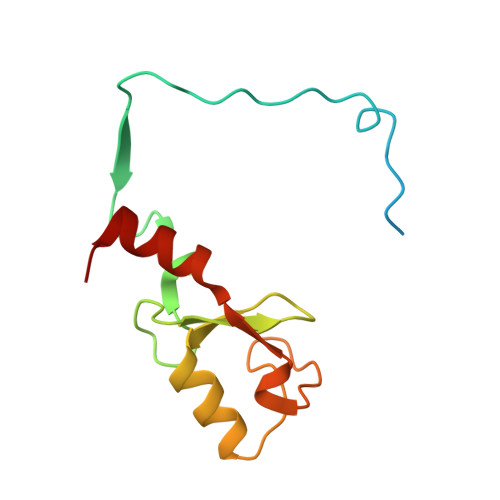
Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex.
Bentley, M.L., Corn, J.E., Dong, K.C., Phung, Q., Cheung, T.K., Cochran, A.G.(2011) EMBO J 30: 3285-3297
- PubMed: 21772249
- DOI: https://doi.org/10.1038/emboj.2011.243
- Primary Citation of Related Structures:
3RPG - PubMed Abstract:
The Polycomb repressive complex 1 (PRC1) mediates gene silencing, in part by monoubiquitination of histone H2A on lysine 119 (uH2A). Bmi1 and Ring1b are critical components of PRC1 that heterodimerize via their N-terminal RING domains to form an active E3 ubiquitin ligase. We have determined the crystal structure of a complex between the Bmi1/Ring1b RING-RING heterodimer and the E2 enzyme UbcH5c and find that UbcH5c interacts with Ring1b only, in a manner fairly typical of E2-E3 interactions. However, we further show that the Bmi1/Ring1b RING domains bind directly to duplex DNA through a basic surface patch unique to the Bmi1/Ring1b RING-RING dimer. Mutation of residues on this interaction surface leads to a loss of H2A ubiquitination activity. Computational modelling of the interface between Bmi1/Ring1b-UbcH5c and the nucleosome suggests that Bmi1/Ring1b interacts with both nucleosomal DNA and an acidic patch on histone H4 to achieve specific monoubiquitination of H2A. Our results point to a novel mechanism of substrate recognition, and control of product formation, by Bmi1/Ring1b.
- Department of Early Discovery Biochemistry, Genentech Research and Early Development, South San Francisco, CA, USA.
Organizational Affiliation: