Structural basis for the recognition of attP substrates by P4-like integrases
Szwagierczak, A., Popowicz, G.M., Holak, T.A., Rakin, A., Antonenka, U.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| CP4-like integrase | A, B [auth C] | 88 | Yersinia pestis | Mutation(s): 0 Gene Names: int, int2, YPO1917, YP_1660, y2393 | 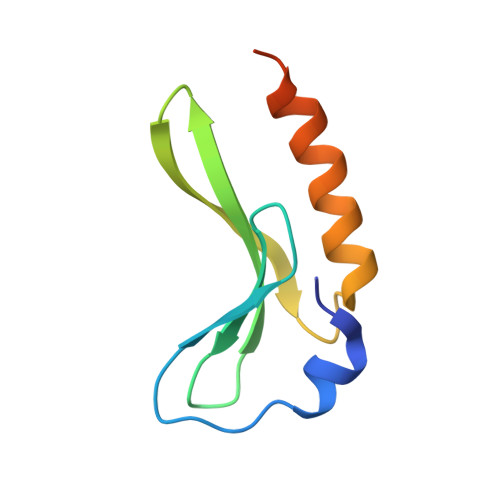 |
UniProt | |||||
Find proteins for Q9Z3B4 (Yersinia pestis) Explore Q9Z3B4 Go to UniProtKB: Q9Z3B4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9Z3B4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 5'-D(*TP*AP*AP*TP*GP*AP*CP*CP*AP*CP*CP*AP*AP*TP*A)-3' | C [auth E], E [auth G] | 15 | N/A | 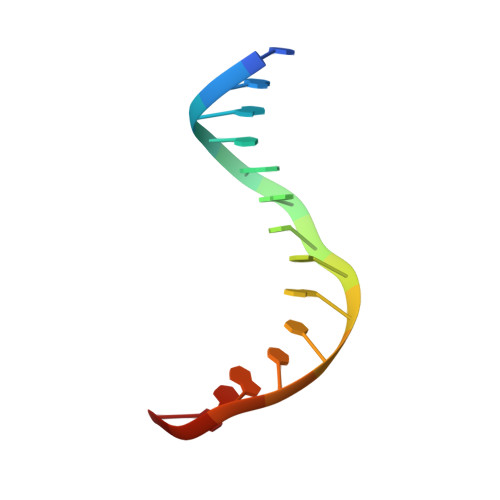 | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 5'-D(*TP*AP*TP*TP*GP*GP*TP*GP*GP*TP*CP*AP*TP*TP*A)-3' | D [auth F], F [auth H] | 15 | N/A | 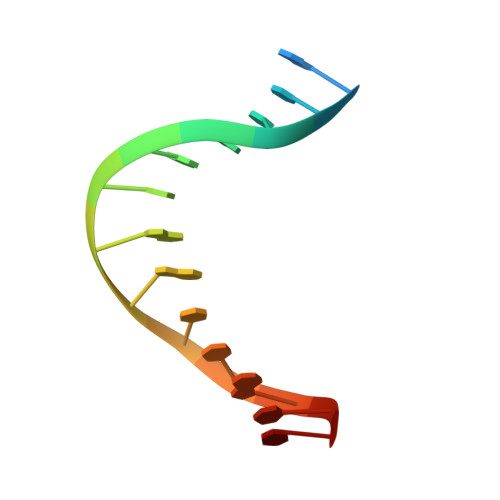 | |
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 60.9 | α = 90 |
| b = 69.49 | β = 90 |
| c = 83.74 | γ = 90 |
| Software Name | Purpose |
|---|---|
| MAR345dtb | data collection |
| MOLREP | phasing |
| REFMAC | refinement |
| XDS | data reduction |
| XSCALE | data scaling |