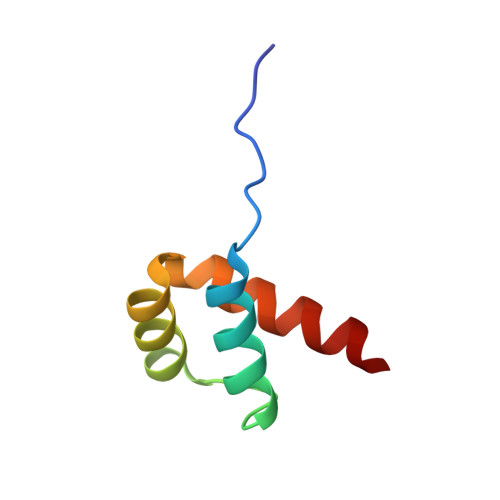
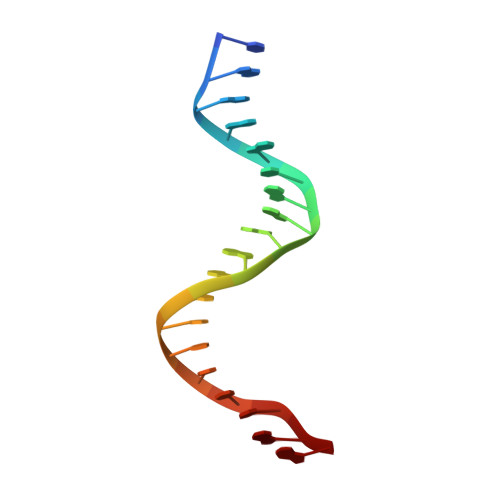
Crystal structure of the human NKX2.5 homeodomain in complex with DNA target.
Pradhan, L., Genis, C., Scone, P., Weinberg, E.O., Kasahara, H., Nam, H.J.(2012) Biochemistry 51: 6312-6319
- PubMed: 22849347
- DOI: https://doi.org/10.1021/bi300849c
- Primary Citation of Related Structures:
3RKQ - PubMed Abstract:
NKX2.5 is a homeodomain containing transcription factor regulating cardiac formation and function, and its mutations are linked to congenital heart disease. Here we provide the first report of the crystal structure of the NKX2.5 homeodomain in complex with double-stranded DNA of its endogenous target, locating within the proximal promoter -242 site of the atrial natriuretic factor gene. The crystal structure, determined at 1.8 Å resolution, demonstrates that NKX2.5 homeodomains occupy both DNA binding sites separated by five nucleotides without physical interaction between themselves. The two homeodomains show identical conformation despite the differences in the DNA sequences they bind, and no significant bending of the DNA was observed. Tyr54, absolutely conserved in NK2 family proteins, mediates sequence-specific interaction with the TAAG motif. This high resolution crystal structure of NKX2.5 protein provides a detailed picture of protein and DNA interactions, which allows us to predict DNA binding of mutants identified in human patients.
- Department of Bioengineering, University of Texas at Dallas, Richardson, TX 75080, USA.
Organizational Affiliation: