Molecular insights into DNA polymerase deterrents for ribonucleotide insertion.
Cavanaugh, N.A., Beard, W.A., Batra, V.K., Perera, L., Pedersen, L.G., Wilson, S.H.(2011) J Biological Chem 286: 31650-31660
- PubMed: 21733843
- DOI: https://doi.org/10.1074/jbc.M111.253401
- Primary Citation of Related Structures:
3RH4, 3RH5, 3RH6 - PubMed Abstract:
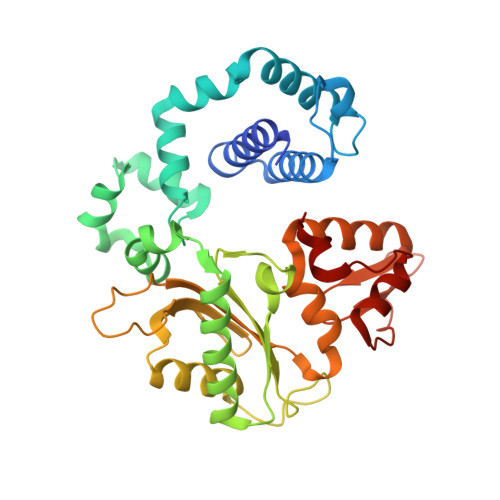
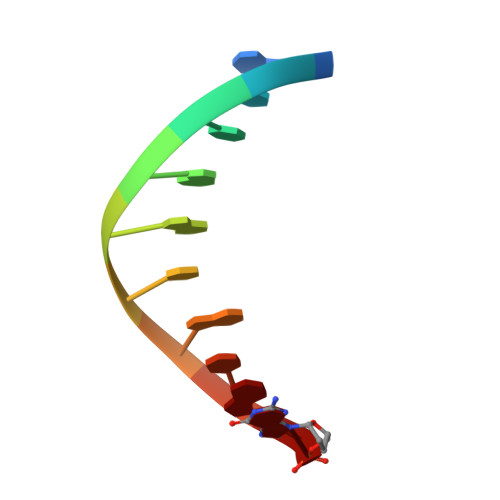
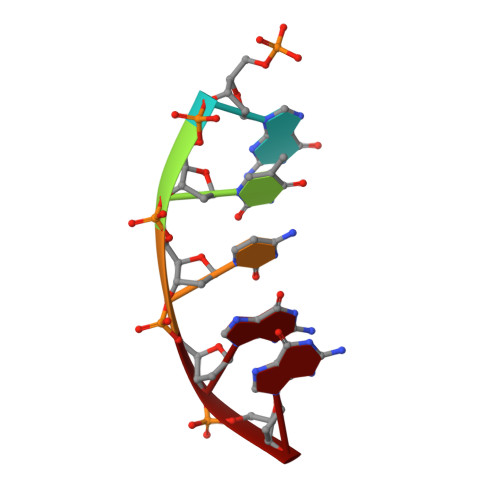
DNA polymerases can misinsert ribonucleotides that lead to genomic instability. DNA polymerase β discourages ribonucleotide insertion with the backbone carbonyl of Tyr-271; alanine substitution of Tyr-271, but not Phe-272, resulted in a >10-fold loss in discrimination. The Y271A mutant also inserted ribonucleotides more efficiently than wild type on a variety of ribonucleoside (rNMP)-containing DNA substrates. Substituting Mn(2+) for Mg(2+) decreased sugar discrimination for both wild-type and mutant enzymes primarily by increasing the affinity for rCTP. This facilitated crystallization of ternary substrate complexes of both the wild-type and Y271A mutant enzymes. Crystallographic structures of Y271A- and wild type-substrate complexes indicated that rCTP is well accommodated in the active site but that O2' of rCTP and the carbonyl oxygen of Tyr-271 or Ala-271 are unusually close (∼2.5 and 2.6 Å, respectively). Structure-based modeling indicates that the local energetic cost of positioning these closely spaced oxygens is ∼2.2 kcal/mol for the wild-type enzyme. Because the side chain of Tyr-271 also hydrogen bonds with the primer terminus, loss of this interaction affects its catalytic positioning. Our results support a model where DNA polymerase β utilizes two strategies, steric and geometric, with a single protein residue to deter ribonucleotide insertion.
- Laboratory of Structural Biology, NIEHS, National Institutes of Health, Research Triangle Park, North Carolina 27709-2233, USA.
Organizational Affiliation: