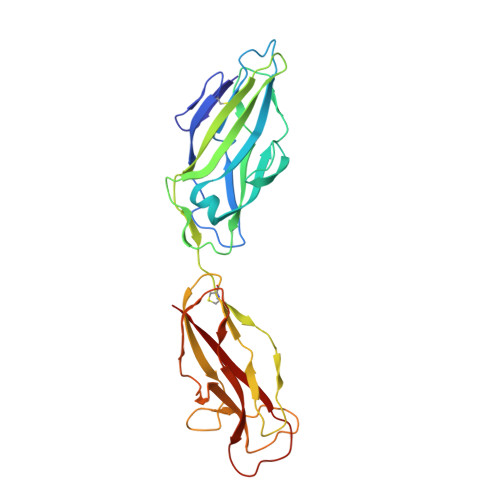
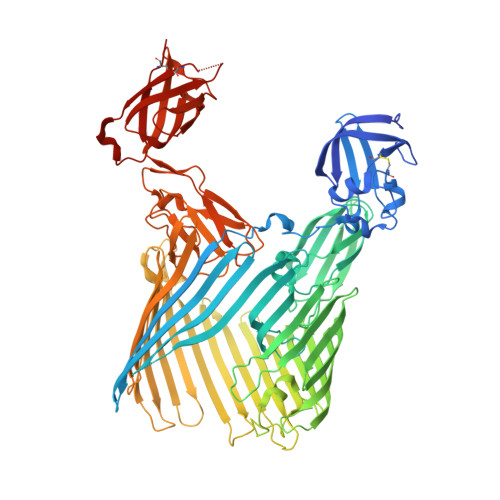
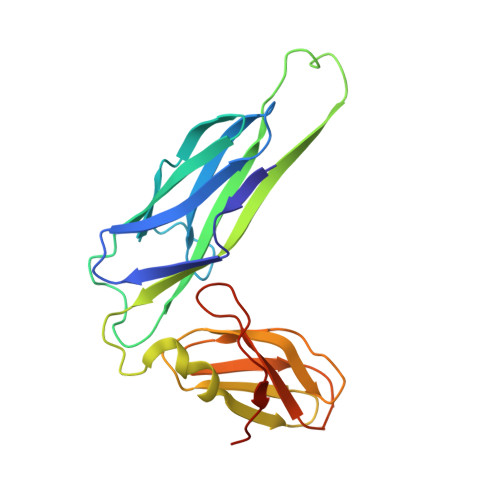
Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate.
Phan, G., Remaut, H., Wang, T., Allen, W.J., Pirker, K.F., Lebedev, A., Henderson, N.S., Geibel, S., Volkan, E., Yan, J., Kunze, M.B., Pinkner, J.S., Ford, B., Kay, C.W., Li, H., Hultgren, S.J., Thanassi, D.G., Waksman, G.(2011) Nature 474: 49-53
- PubMed: 21637253
- DOI: https://doi.org/10.1038/nature10109
- Primary Citation of Related Structures:
3OHN, 3RFZ - PubMed Abstract:
Type 1 pili are the archetypal representative of a widespread class of adhesive multisubunit fibres in Gram-negative bacteria. During pilus assembly, subunits dock as chaperone-bound complexes to an usher, which catalyses their polymerization and mediates pilus translocation across the outer membrane. Here we report the crystal structure of the full-length FimD usher bound to the FimC-FimH chaperone-adhesin complex and that of the unbound form of the FimD translocation domain. The FimD-FimC-FimH structure shows FimH inserted inside the FimD 24-stranded β-barrel translocation channel. FimC-FimH is held in place through interactions with the two carboxy-terminal periplasmic domains of FimD, a binding mode confirmed in solution by electron paramagnetic resonance spectroscopy. To accommodate FimH, the usher plug domain is displaced from the barrel lumen to the periplasm, concomitant with a marked conformational change in the β-barrel. The amino-terminal domain of FimD is observed in an ideal position to catalyse incorporation of a newly recruited chaperone-subunit complex. The FimD-FimC-FimH structure provides unique insights into the pilus subunit incorporation cycle, and captures the first view of a protein transporter in the act of secreting its cognate substrate.
- Institute of Structural and Molecular Biology, University College London and Birkbeck College, Malet Street, London WC1E 7HX, UK.
Organizational Affiliation: