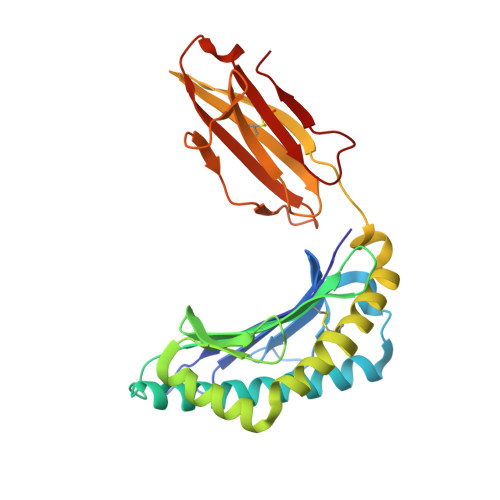
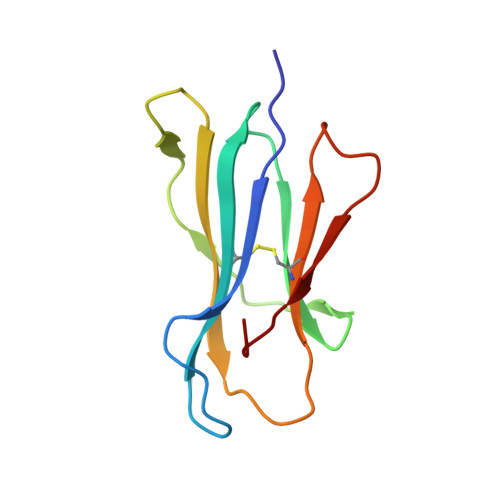
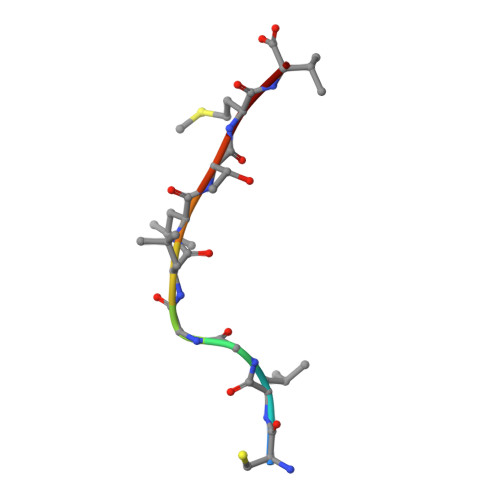
Structural and energetic evidence for highly peptide-specific tumor antigen targeting via allo-MHC restriction.
Simpson, A.A., Mohammed, F., Salim, M., Tranter, A., Rickinson, A.B., Stauss, H.J., Moss, P.A., Steven, N.M., Willcox, B.E.(2011) Proc Natl Acad Sci U S A 108: 21176-21181
- PubMed: 22160697
- DOI: https://doi.org/10.1073/pnas.1108422109
- Primary Citation of Related Structures:
3REV, 3REW - PubMed Abstract:
Immunotherapies targeting peptides presented by allogeneic MHC molecules offer the prospect of circumventing tolerance to key tumor-associated self-antigens. However, the degree of antigen specificity mediated by alloreactive T cells, and their ability to discriminate normal tissues from transformed cells presenting elevated antigen levels, is poorly understood. We examined allorecognition of an HLA-A2-restricted Hodgkin's lymphoma-associated antigen and were able to isolate functionally antigen-specific allo-HLA-A2-restricted T cells from multiple donors. Binding and structural studies, focused on a prototypic allo-HLA-A2-restricted T-cell receptor (TCR) termed NB20 derived from an HLA-A3 homozygote, suggested highly peptide-specific allorecognition that was energetically focused on antigen, involving direct recognition of a distinct allopeptide presented within a conserved MHC recognition surface. Although NB20/HLA-A2 affinity was unremarkable, TCR/MHC complexes were very short-lived, consistent with suboptimal TCR triggering and tolerance to low antigen levels. These data provide strong molecular evidence that within the functionally heterogeneous alloreactive repertoire, there is the potential for highly antigen-specific "allo-MHC-restricted" recognition and suggest a kinetic mechanism whereby allo-MHC-restricted T cells may discriminate normal from transformed tissue, thereby outlining a suitable basis for broad-based therapeutic targeting of tolerizing tumor antigens.
- Birmingham Cancer Research UK Cancer Centre, School of Cancer Sciences, University of Birmingham, Birmingham B15 2TT, United Kingdom.
Organizational Affiliation: