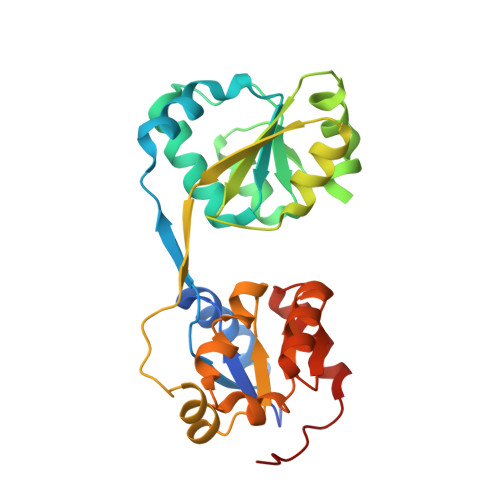
Crystal structure of uroporphyrinogen III synthase from Pseudomonas syringae pv. tomato DC3000
Peng, S.X., Zhang, H., Gao, Y., Pan, X., Cao, P., Li, M., Chang, W.R.(2011) Biochem Biophys Res Commun 408: 576-581
- PubMed: 21527255
- DOI: https://doi.org/10.1016/j.bbrc.2011.04.064
- Primary Citation of Related Structures:
3RE1 - PubMed Abstract:
Uroporphyrinogen III synthase (U3S) is one of the key enzymes in the biosynthesis of tetrapyrrole compounds. It catalyzes the cyclization of the linear hydroxymethylbilane (HMB) to uroporphyrinogen III (uro'gen III). We have determined the crystal structure of U3S from Pseudomonas syringae pv. tomato DC3000 (psU3S) at 2.5Å resolution by the single wavelength anomalous dispersion (SAD) method. Each psU3S molecule consists of two domains interlinked by a two-stranded antiparallel β-sheet. The conformation of psU3S is different from its homologous proteins because of the flexibility of the linker between the two domains, which might be related to this enzyme's catalytic properties. Based on mutation and activity analysis, a key residue, Arg219, was found to be important for the catalytic activity of psU3S. Mutation of Arg219 to Ala caused a decrease in enzymatic activity to about 25% that of the wild type enzyme. Our results provide the structural basis and biochemical evidence to further elucidate the catalytic mechanism of U3S.
- National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, 15 Datun Road, Chaoyang District, Beijing, PR China.
Organizational Affiliation: