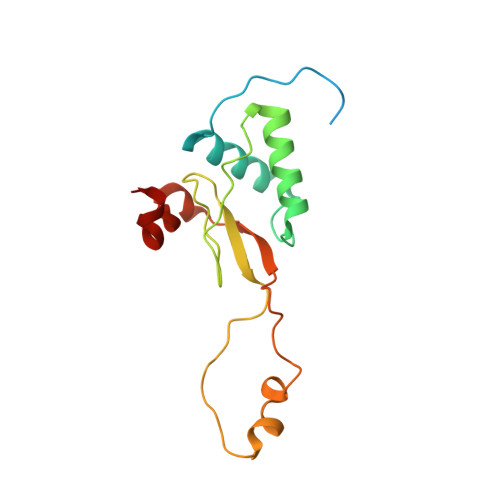
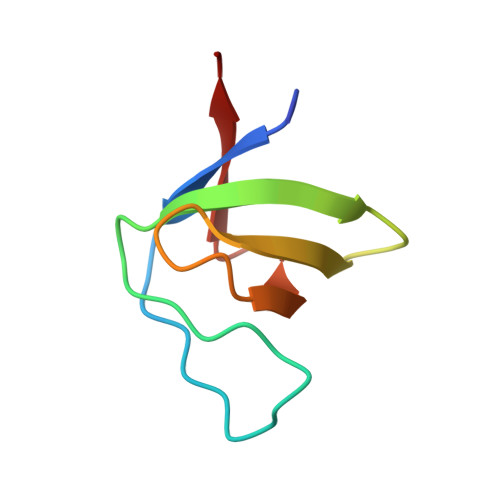
Conformation of the Dileucine-Based Sorting Motif in HIV-1 Nef Revealed by Intermolecular Domain Assembly.
Horenkamp, F.A., Breuer, S., Schulte, A., Lulf, S., Weyand, M., Saksela, K., Geyer, M.(2011) Traffic 12: 867-877
- PubMed: 21477083
- DOI: https://doi.org/10.1111/j.1600-0854.2011.01205.x
- Primary Citation of Related Structures:
3RBB - PubMed Abstract:
The human immunodeficiency virus 1 (HIV-1) Nef protein is a pathogenicity factor required for effective progression to AIDS, which modulates host cell signaling pathways and T-cell receptor internalization. We have determined the crystal structure of Nef, allele SF2, in complex with an engineered SH3 domain of human Hck showing unnaturally tight binding and inhibitory potential toward Nef. This complex provides the most complete Nef structure described today, and explains the structural basis of the high affinity of this interaction. Intriguingly, the 33-residue C-terminal flexible loop is resolved in the structure by its interactions with a highly conserved hydrophobic groove on the core domain of an adjacent Nef molecule. The loop mediates the interaction of Nef with the cellular adaptor protein machinery for the stimulated internalization of surface receptors. The endocytic dileucine-based sorting motif is exposed at the tip of the acidic loop, giving the myristoylated Nef protein a distinctly dipolar character. The intermolecular domain assembly of Nef provides insights into a possible regulation mechanism for cargo trafficking.
- Abteilung Physikalische Biochemie, Max-Planck-Institut für molekulare Physiologie, Otto-Hahn-Strasse 11, 44227 Dortmund, Germany.
Organizational Affiliation: