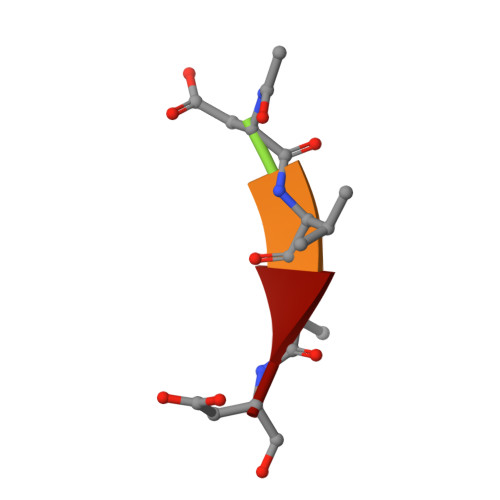
Structural and enzymatic insights into caspase-2 protein substrate recognition and catalysis.
Tang, Y., Wells, J.A., Arkin, M.R.(2011) J Biological Chem 286: 34147-34154
- PubMed: 21828056
- DOI: https://doi.org/10.1074/jbc.M111.247627
- Primary Citation of Related Structures:
3R5J, 3R6G, 3R6L, 3R7B, 3R7N, 3R7S - PubMed Abstract:
Caspase-2, the most evolutionarily conserved member in the human caspase family, may play important roles in stress-induced apoptosis, cell cycle regulation, and tumor suppression. In biochemical assays, caspase-2 uniquely prefers a pentapeptide (such as VDVAD) rather than a tetrapeptide, as required for efficient cleavage by other caspases. We investigated the molecular basis for pentapeptide specificity using peptide analog inhibitors and substrates that vary at the P5 position. We determined the crystal structures of apo caspase-2, caspase-2 in complex with peptide inhibitors VDVAD-CHO, ADVAD-CHO, and DVAD-CHO, and a T380A mutant of caspase-2 in complex with VDVAD-CHO. Two residues, Thr-380 and Tyr-420, are identified to be critical for the P5 residue recognition; mutation of the two residues reduces the catalytic efficiency by about 4- and 40-fold, respectively. The structures also provide a series of snapshots of caspase-2 in different catalytic states, shedding light on the mechanism of capase-2 activation, substrate binding, and catalysis. By comparing the apo and inhibited caspase-2 structures, we propose that the disruption of a non-conserved salt bridge between Glu-217 and the invariant Arg-378 is important for the activation of caspase-2. These findings broaden our understanding of caspase-2 substrate specificity and catalysis.
- Department of Pharmaceutical Chemistry, Small Molecule Discovery Center, University of California, San Francisco, California 94158, USA.
Organizational Affiliation: