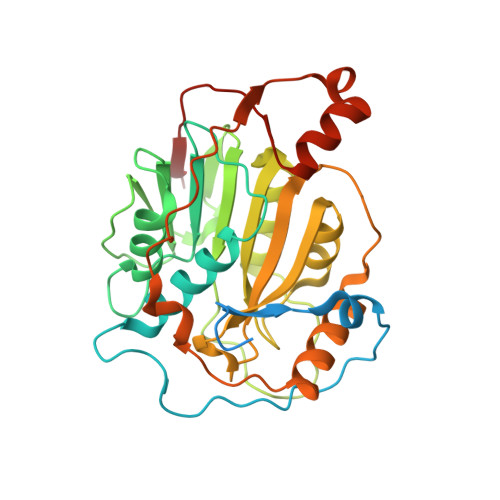
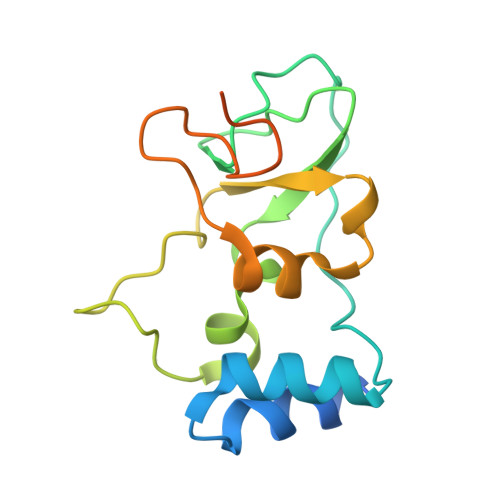
Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex.
Chen, Y., Su, C., Ke, M., Jin, X., Xu, L., Zhang, Z., Wu, A., Sun, Y., Yang, Z., Tien, P., Ahola, T., Liang, Y., Liu, X., Guo, D.(2011) PLoS Pathog 7: e1002294-e1002294
- PubMed: 22022266
- DOI: https://doi.org/10.1371/journal.ppat.1002294
- Primary Citation of Related Structures:
3R24 - PubMed Abstract:
The 5'-cap structure is a distinct feature of eukaryotic mRNAs, and eukaryotic viruses generally modify the 5'-end of viral RNAs to mimic cellular mRNA structure, which is important for RNA stability, protein translation and viral immune escape. SARS coronavirus (SARS-CoV) encodes two S-adenosyl-L-methionine (SAM)-dependent methyltransferases (MTase) which sequentially methylate the RNA cap at guanosine-N7 and ribose 2'-O positions, catalyzed by nsp14 N7-MTase and nsp16 2'-O-MTase, respectively. A unique feature for SARS-CoV is that nsp16 requires non-structural protein nsp10 as a stimulatory factor to execute its MTase activity. Here we report the biochemical characterization of SARS-CoV 2'-O-MTase and the crystal structure of nsp16/nsp10 complex bound with methyl donor SAM. We found that SARS-CoV nsp16 MTase methylated m7GpppA-RNA but not m7GpppG-RNA, which is in contrast with nsp14 MTase that functions in a sequence-independent manner. We demonstrated that nsp10 is required for nsp16 to bind both m7GpppA-RNA substrate and SAM cofactor. Structural analysis revealed that nsp16 possesses the canonical scaffold of MTase and associates with nsp10 at 1∶1 ratio. The structure of the nsp16/nsp10 interaction interface shows that nsp10 may stabilize the SAM-binding pocket and extend the substrate RNA-binding groove of nsp16, consistent with the findings in biochemical assays. These results suggest that nsp16/nsp10 interface may represent a better drug target than the viral MTase active site for developing highly specific anti-coronavirus drugs.
- State Key Laboratory of Virology and Modern Virology Research Center, College of Life Sciences, Wuhan University, Wuhan, P. R. China.
Organizational Affiliation: