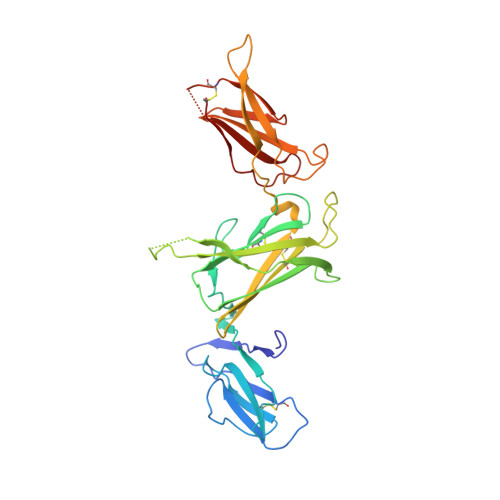
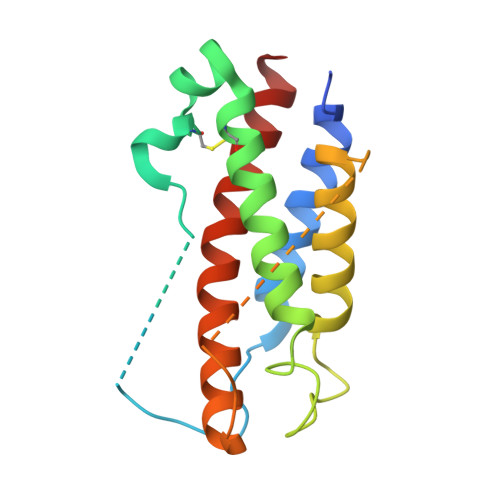
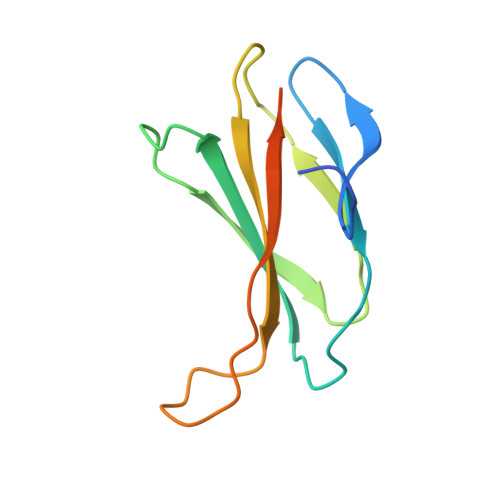
Structures of adnectin/protein complexes reveal an expanded binding footprint.
Ramamurthy, V., Krystek, S.R., Bush, A., Wei, A., Emanuel, S.L., Das Gupta, R., Janjua, A., Cheng, L., Murdock, M., Abramczyk, B., Cohen, D., Lin, Z., Morin, P., Davis, J.H., Dabritz, M., McLaughlin, D.C., Russo, K.A., Chao, G., Wright, M.C., Jenny, V.A., Engle, L.J., Furfine, E., Sheriff, S.(2012) Structure 20: 259-269
- PubMed: 22325775
- DOI: https://doi.org/10.1016/j.str.2011.11.016
- Primary Citation of Related Structures:
3QWQ, 3QWR - PubMed Abstract:
Adnectins are targeted biologics derived from the tenth type III domain of human fibronectin (¹⁰Fn3), a member of the immunoglobulin superfamily. Target-specific binders are selected from libraries generated by diversifying the three ¹⁰Fn3 loops that are analogous to the complementarity determining regions of antibodies. The crystal structures of two Adnectins were determined, each in complex with its therapeutic target, EGFR or IL-23. Both Adnectins bind different epitopes than those bound by known monoclonal antibodies. Molecular modeling suggests that some of these epitopes might not be accessible to antibodies because of the size and concave shape of the antibody combining site. In addition to interactions from the Adnectin diversified loops, residues from the N terminus and/or the β strands interact with the target proteins in both complexes. Alanine-scanning mutagenesis confirmed the calculated binding energies of these β strand interactions, indicating that these nonloop residues can expand the available binding footprint.
- Bristol-Myers Squibb Research & Development, Princeton, NJ 08543-4000, USA.
Organizational Affiliation: