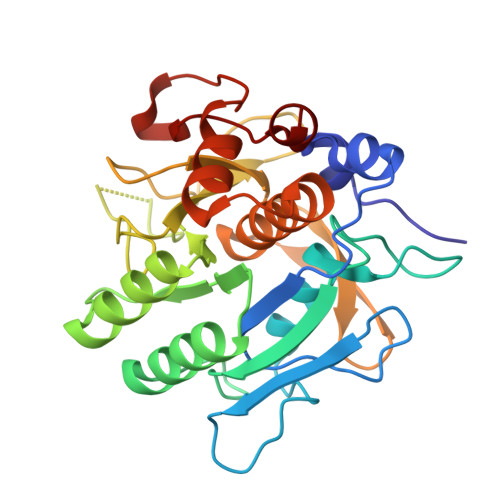
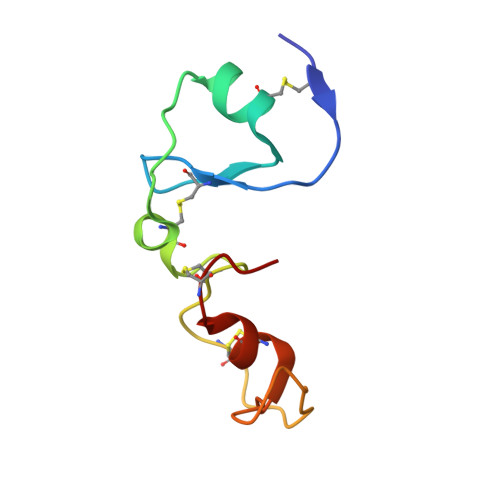
Structural basis for dual-inhibition mechanism of a non-classical kazal-type serine protease inhibitor from horseshoe crab in complex with subtilisin.
Shenoy, R.T., Thangamani, S., Velazquez-Campoy, A., Ho, B., Ding, J.L., Sivaraman, J.(2011) PLoS One 6: e18838-e18838
- PubMed: 21541315
- DOI: https://doi.org/10.1371/journal.pone.0018838
- Primary Citation of Related Structures:
3QTL - PubMed Abstract:
Serine proteases play a crucial role in host-pathogen interactions. In the innate immune system of invertebrates, multi-domain protease inhibitors are important for the regulation of host-pathogen interactions and antimicrobial activities. Serine protease inhibitors, 9.3-kDa CrSPI isoforms 1 and 2, have been identified from the hepatopancreas of the horseshoe crab, Carcinoscorpius rotundicauda. The CrSPIs were biochemically active, especially CrSPI-1, which potently inhibited subtilisin (Ki = 1.43 nM). CrSPI has been grouped with the non-classical Kazal-type inhibitors due to its unusual cysteine distribution. Here we report the crystal structure of CrSPI-1 in complex with subtilisin at 2.6 Å resolution and the results of biophysical interaction studies. The CrSPI-1 molecule has two domains arranged in an extended conformation. These two domains act as heads that independently interact with two separate subtilisin molecules, resulting in the inhibition of subtilisin activity at a ratio of 1:2 (inhibitor to protease). Each subtilisin molecule interacts with the reactive site loop from each domain of CrSPI-1 through a standard canonical binding mode and forms a single ternary complex. In addition, we propose the substrate preferences of each domain of CrSPI-1. Domain 2 is specific towards the bacterial protease subtilisin, while domain 1 is likely to interact with the host protease, Furin. Elucidation of the structure of the CrSPI-1: subtilisin (1∶2) ternary complex increases our understanding of host-pathogen interactions in the innate immune system at the molecular level and provides new strategies for immunomodulation.
- Department of Biological Sciences, National University of Singapore, Singapore, Singapore.
Organizational Affiliation: