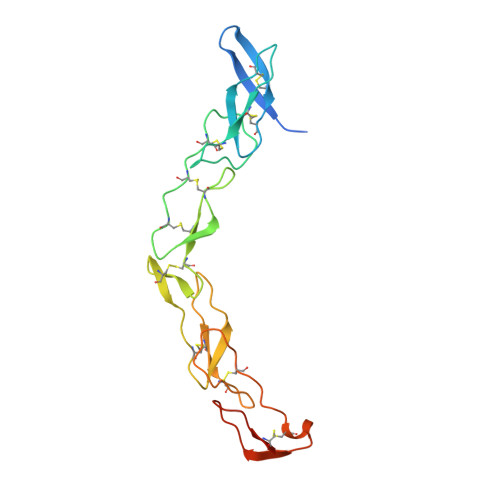
The crystal structure of death receptor 6 (DR6): a potential receptor of the amyloid precursor protein (APP).
Kuester, M., Kemmerzehl, S., Dahms, S.O., Roeser, D., Than, M.E.(2011) J Mol Biology 409: 189-201
- PubMed: 21463639
- DOI: https://doi.org/10.1016/j.jmb.2011.03.048
- Primary Citation of Related Structures:
3QO4 - PubMed Abstract:
Death receptors belong to the tumor necrosis factor receptor (TNFR) super family and are intimately involved in the signal transduction during apoptosis, stress response and cellular survival. Here we present the crystal structure of recombinantly expressed death receptor six (DR6), one family member that was recently shown to bind to the amyloid precursor protein (APP) and hence to be probably involved in the development of Alzheimer's disease. The extracellular cysteine rich region of DR6, the typical ligand binding region of all TNFRs, was refined to 2.2 Å resolution and shows that its four constituting cysteine rich domains (CRDs) are arranged in a rod-like overall structure, which presents DR6-specific surface patches responsible for the exclusive recognition of its ligand(s). Based on the structural data, the general ligand binding modes of TNFRs and molecular modeling experiments we were able to elucidate structural features of the potential DR6-APP signaling complex.
- Leibniz Institute for Age Research-Fritz Lipmann Institut, Protein Crystallography Group, Beutenbergstr. 11, 07745 Jena, Germany.
Organizational Affiliation: