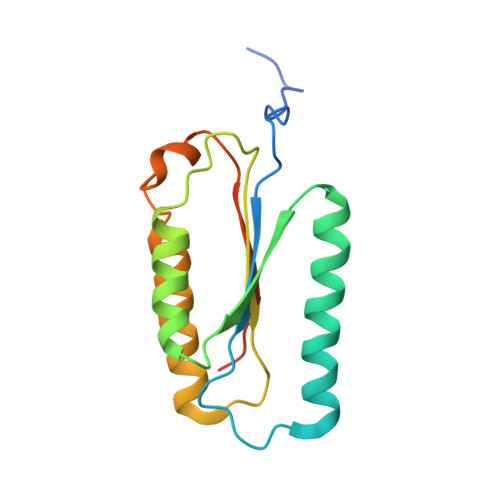
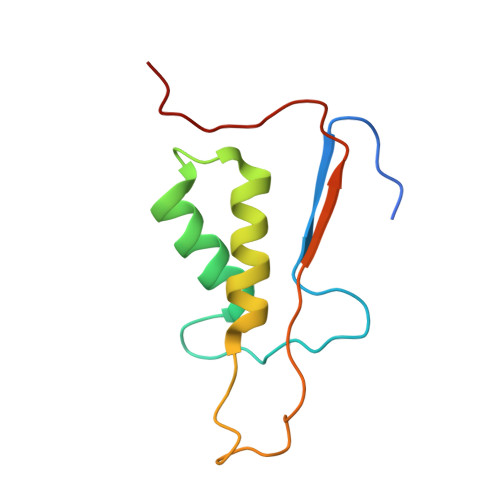
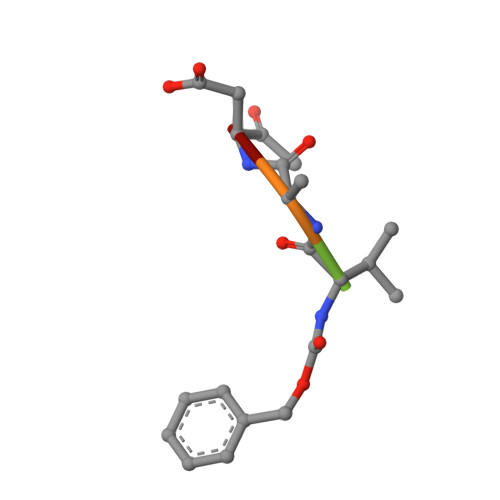
Structure of human caspase-6 in complex with Z-VAD-FMK: New peptide binding mode observed for the non-canonical caspase conformation.
Muller, I., Lamers, M.B., Ritchie, A.J., Dominguez, C., Munoz-Sanjuan, I., Kiselyov, A.(2011) Bioorg Med Chem Lett 21: 5244-5247
- PubMed: 21820899
- DOI: https://doi.org/10.1016/j.bmcl.2011.07.041
- Primary Citation of Related Structures:
3QNW - PubMed Abstract:
Caspase-6 is a cysteine protease implicated in neuronal survival and apoptosis. Deregulation of caspase-6 activity was linked to several neurodegenerative disorders including Alzheimer's and Huntington's Diseases. Several recent studies on the structure of caspase-6 feature the caspase-6 zymogen, mature apo-caspase-6 as well as the Ac-VEID-CHO peptide complex. All structures share the same typical dimeric caspase conformation. However, mature apo-caspase-6 crystallized at low pH revealed a novel, non-canonical inactive caspase conformation speculated to represent a latent state of the enzyme suitable for the design of allosteric inhibitors. In this treatise we present the structure of caspase-6 in the non-canonical inactive enzyme conformation bound to the irreversible inhibitor Z-VAD-FMK. The complex features a unique peptide binding mode not observed previously.
- BioFocus, Chesterford Research Park, Saffron Walden, Essex, CB10 1XL, UK.
Organizational Affiliation: