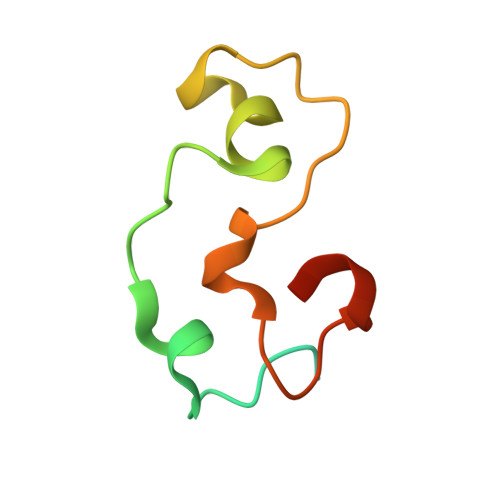
The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain.
Xu, C., Bian, C., Lam, R., Dong, A., Min, J.(2011) Nat Commun 2: 227-227
- PubMed: 21407193
- DOI: https://doi.org/10.1038/ncomms1237
- Primary Citation of Related Structures:
3QMB, 3QMC, 3QMD, 3QMG, 3QMH, 3QMI - PubMed Abstract:
CFP1 is a CXXC domain-containing protein and an essential component of the SETD1 histone H3K4 methyltransferase complex. CXXC domain proteins direct different chromatin-modifying activities to various chromatin regions. Here, we report crystal structures of the CFP1 CXXC domain in complex with six different CpG DNA sequences. The crescent-shaped CFP1 CXXC domain is wedged into the major groove of the CpG DNA, distorting the B-form DNA, and interacts extensively with the major groove of the DNA. The structures elucidate the molecular mechanism of the non-methylated CpG-binding specificity of the CFP1 CXXC domain. The CpG motif is confined by a tripeptide located in a rigid loop, which only allows the accommodation of the non-methylated CpG dinucleotide. Furthermore, we demonstrate that CFP1 has a preference for a guanosine nucleotide following the CpG motif.
- Structural Genomics Consortium, University of Toronto, Toronto, Ontario, Canada.
Organizational Affiliation: