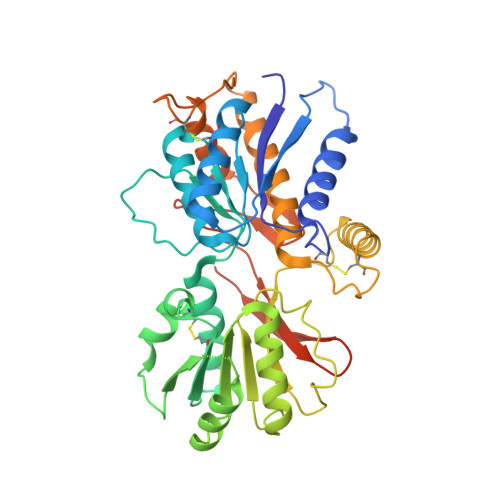
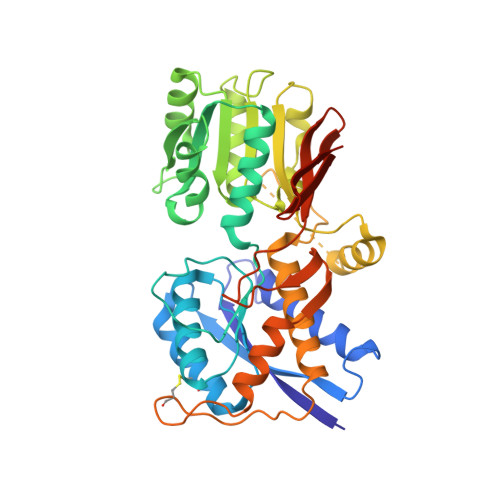
Structure and assembly mechanism for heteromeric kainate receptors.
Kumar, J., Schuck, P., Mayer, M.L.(2011) Neuron 71: 319-331
- PubMed: 21791290
- DOI: https://doi.org/10.1016/j.neuron.2011.05.038
- Primary Citation of Related Structures:
3QLT, 3QLU, 3QLV - PubMed Abstract:
Native glutamate receptor ion channels are tetrameric assemblies containing two or more different subunits. NMDA receptors are obligate heteromers formed by coassembly of two or three divergent gene families. While some AMPA and kainate receptors can form functional homomeric ion channels, the KA1 and KA2 subunits are obligate heteromers which function only in combination with GluR5-7. The mechanisms controlling glutamate receptor assembly involve an initial step in which the amino terminal domains (ATD) assemble as dimers. Here, we establish by sedimentation velocity that the ATDs of GluR6 and KA2 coassemble as a heterodimer of K(d) 11 nM, 32,000-fold lower than the K(d) for homodimer formation by KA2; we solve crystal structures for the GluR6/KA2 ATD heterodimer and heterotetramer assemblies. Using these structures as a guide, we perform a mutant cycle analysis to probe the energetics of assembly and show that high-affinity ATD interactions are required for biosynthesis of functional heteromeric receptors.
- Laboratory of Cellular and Molecular Neurophysiology, Porter Neuroscience Research Center, NICHD, NIH, DHHS, Bethesda, MD 20892, USA.
Organizational Affiliation: