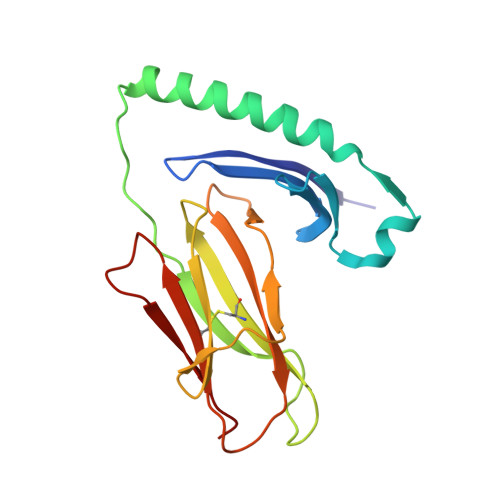
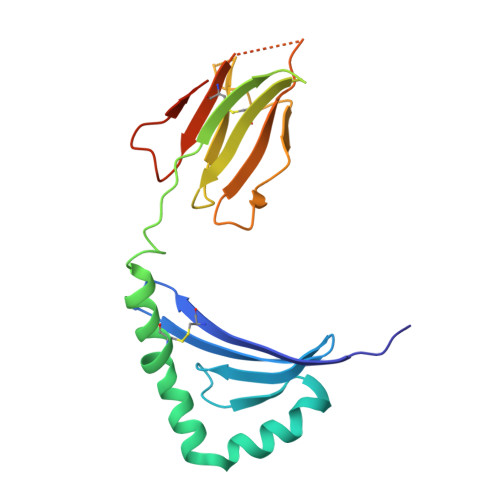
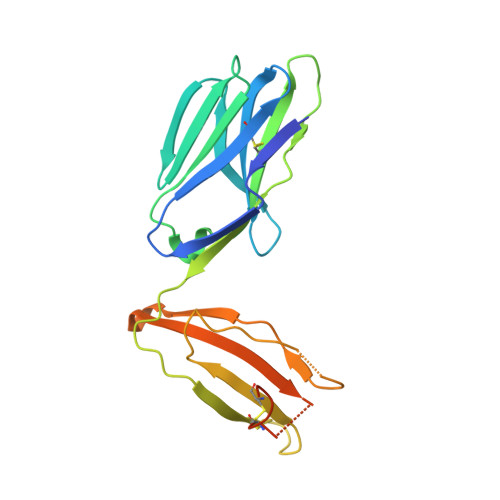
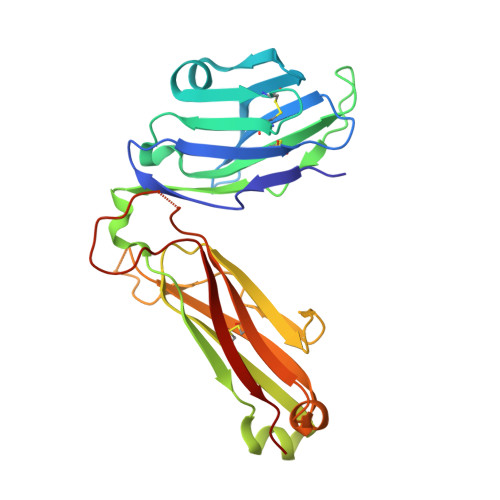
Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k).
Newell, E.W., Ely, L.K., Kruse, A.C., Reay, P.A., Rodriguez, S.N., Lin, A.E., Kuhns, M.S., Garcia, K.C., Davis, M.M.(2011) J Immunol 186: 5823-5832
- PubMed: 21490152
- DOI: https://doi.org/10.4049/jimmunol.1100197
- Primary Citation of Related Structures:
3QIB, 3QIU, 3QIW, 3QJF, 3QJH - PubMed Abstract:
T cells specific for the cytochrome c Ag are widely used to investigate many aspects of TCR specificity and interactions with peptide-MHC, but structural information has long been elusive. In this study, we present structures for the well-studied 2B4 TCR, as well as a naturally occurring variant of the 5c.c7 TCR, 226, which is cross-reactive with more than half of possible substitutions at all three TCR-sensitive residues on the peptide Ag. These structures alone and in complex with peptide-MHC ligands allow us to reassess many prior mutagenesis results. In addition, the structure of 226 bound to one peptide variant, p5E, shows major changes in the CDR3 contacts compared with wild-type, yet the TCR V-region contacts with MHC are conserved. These and other data illustrate the ability of TCRs to accommodate large variations in CDR3 structure and peptide contacts within the constraints of highly conserved TCR-MHC interactions.
- Department of Microbiology and Immunology, Stanford University School of Medicine, Stanford University, Stanford, CA 94305, USA.
Organizational Affiliation: