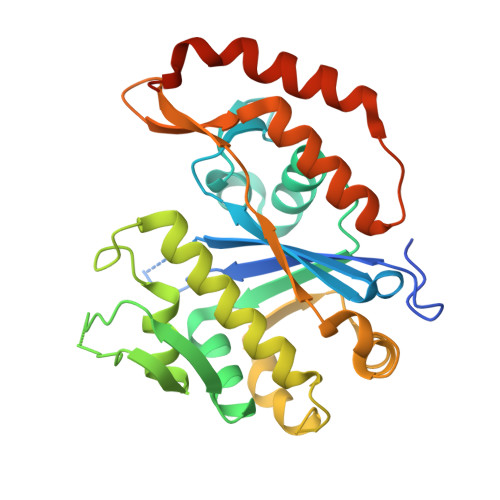
X-ray crystal structure of transcriptional regulator (EF0465) from Enterococcus faecalis, Northeast Structural Genomics Consortium Target EfR190
Forouhar, F., Neely, H., Seetharaman, J., Wang, H., Ciccosanti, C., Mao, L., Acton, T.B., Xiao, R., Everett, J.K., Montelione, G.T., Tong, L., Hun, J.F., Northeast Structural Genomics Consortium (NESG)To be published.