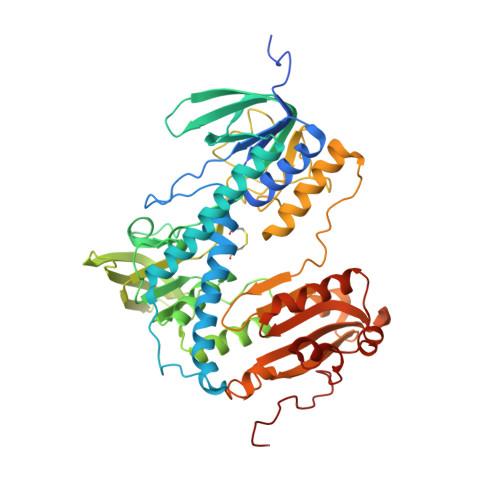
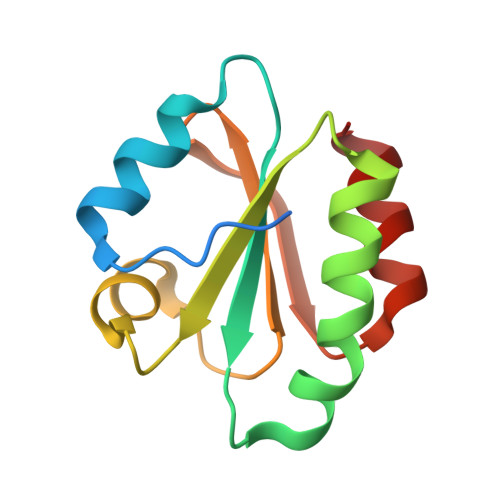
Crystal structure of the human thioredoxin reductase-thioredoxin complex
Fritz-Wolf, K., Kehr, S., Stumpf, M., Rahlfs, S., Becker, K.(2011) Nat Commun 2: 383-383
- PubMed: 21750537
- DOI: https://doi.org/10.1038/ncomms1382
- Primary Citation of Related Structures:
3QFA, 3QFB - PubMed Abstract:
Thioredoxin reductase 1 (TrxR1) is a homodimeric flavoprotein crucially involved in the regulation of cellular redox homeostasis, growth, and differentiation. Its importance in various diseases makes TrxR1 a highly interesting drug target. Here we present the first crystal structures of human TrxR1 in complex with its substrate thioredoxin (Trx). The carboxy-terminal redox centre is found about 20 Å apart from the amino-terminal redox centre, with no major conformational changes in TrxR or Trx. Thus, our structure confirms that the enzyme uses a flexible C-terminal arm for electron transport to its substrates, which is stabilized by a guiding bar for controlled transfer. This notion is supported by mutational analyses. Furthermore, essential residues of the interface region were characterized both structurally and functionally. The structure provides templates for future drug design, and contributes to our understanding of redox regulatory processes in mammals.
- Interdisciplinary Research Centre, Justus Liebig University, D-35392 Giessen, Germany.
Organizational Affiliation: