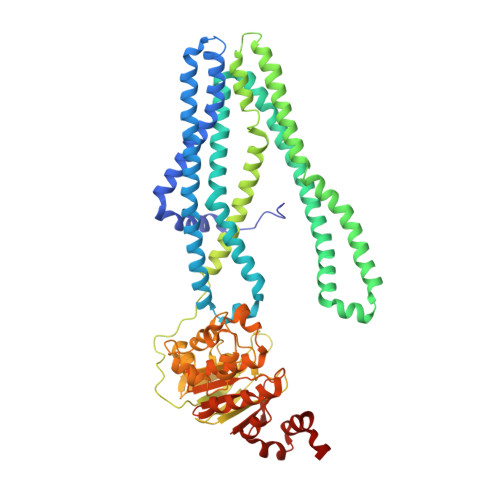
Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation
Hohl, M., Briand, C., Grutter, M.G., Seeger, M.A.(2012) Nat Struct Mol Biol 19: 395-402
- PubMed: 22447242
- DOI: https://doi.org/10.1038/nsmb.2267
- Primary Citation of Related Structures:
3QF4 - PubMed Abstract:
ATP-binding cassette (ABC) transporters shuttle a wide variety of molecules across cell membranes by alternating between inward- and outward-facing conformations, harnessing the energy of ATP binding and hydrolysis at their nucleotide binding domains (NBDs). Here we present the 2.9-Å crystal structure of the heterodimeric ABC transporter TM287-TM288 (TM287/288) from Thermotoga maritima in its inward-facing state. In contrast to previous studies, we found that the NBDs only partially separate, remaining in contact through an interface involving conserved motifs that connect the two ATP hydrolysis sites. We observed AMP-PNP binding to the degenerate catalytic site, which deviates from the consensus sequence in the same positions as the eukaryotic homologs CFTR and TAP1-TAP2 (TAP1/2). The TM287/288 structure provides unprecedented insights into the mechanism of heterodimeric ABC exporters and will enable future studies on this large transporter superfamily.
- Department of Biochemistry, University of Zürich, Zürich, Switzerland.
Organizational Affiliation: