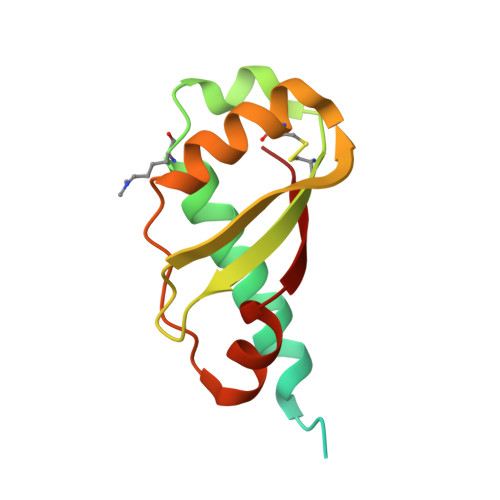
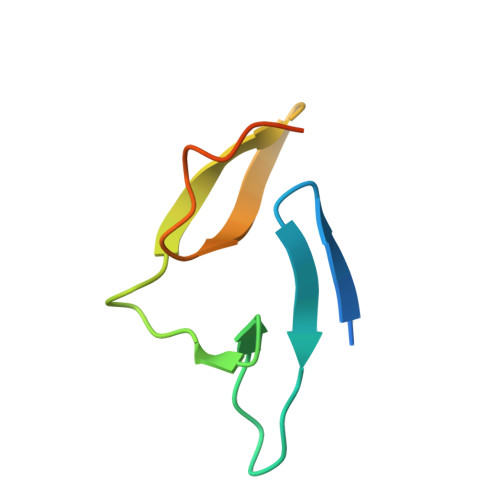
Structural Evidence That Colicin A Protein Binds to a Novel Binding Site of TolA Protein in Escherichia coli Periplasm.
Li, C., Zhang, Y., Vankemmelbeke, M., Hecht, O., Aleanizy, F.S., Macdonald, C., Moore, G.R., James, R., Penfold, C.N.(2012) J Biological Chem 287: 19048-19057
- PubMed: 22493500
- DOI: https://doi.org/10.1074/jbc.M112.342246
- Primary Citation of Related Structures:
3QDP, 3QDR - PubMed Abstract:
The Tol assembly of proteins is an interacting network of proteins located in the Escherichia coli cell envelope that transduces energy and contributes to cell integrity. TolA is central to this network linking the inner and outer membranes by interactions with TolQ, TolR, TolB, and Pal. Group A colicins, such as ColA, parasitize the Tol network through interactions with TolA and/or TolB to facilitate translocation through the cell envelope to reach their cytotoxic site of action. We have determined the first structure of the C-terminal domain of TolA (TolAIII) bound to an N-terminal ColA polypeptide (TA(53-107)). The interface region of the TA(53-107)-TolAIII complex consists of polar contacts linking residues Arg-92 to Arg-96 of ColA with residues Leu-375-Pro-380 of TolA, which constitutes a β-strand addition commonly seen in more promiscuous protein-protein contacts. The interface region also includes three cation-π interactions (Tyr-58-Lys-368, Tyr-90-Lys-379, Phe-94-Lys-396), which have not been observed in any other colicin-Tol protein complex. Mutagenesis of the interface residues of ColA or TolA revealed that the effect on the interaction was cumulative; single mutations of either partner had no effect on ColA activity, whereas mutations of three or more residues significantly reduced ColA activity. Mutagenesis of the aromatic ring component of the cation-π interacting residues showed Tyr-58 of ColA to be essential for the stability of complex formation. TA(53-107) binds on the opposite side of TolAIII to that used by g3p, ColN, or TolB, illustrating the flexible nature of TolA as a periplasmic hub protein.
- School of Molecular Medical Sciences, Centre for Biomolecular Sciences, University of Nottingham, University Park, Nottingham NG7 2RD, United Kingdom.
Organizational Affiliation: