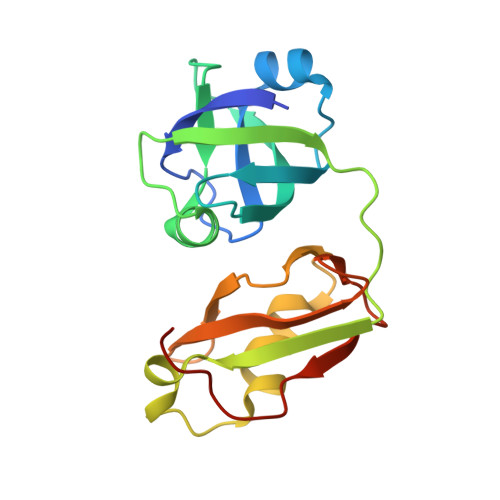
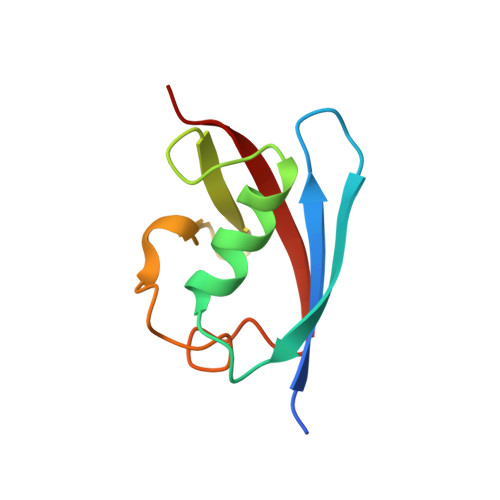
Crystal structure of FAF1 UBX domain in complex with p97/VCP N domain reveals a conformational change in the conserved FcisP touch-turn motif of UBX domain
Kim, K.H., Kang, W., Suh, S.W., Yang, J.K.(2011) Proteins 79: 2583-2587
- PubMed: 21739474
- DOI: https://doi.org/10.1002/prot.23073
- Primary Citation of Related Structures:
3QC8 - Department of Chemistry, College of Natural Sciences, Seoul National University, Seoul, Korea.
Organizational Affiliation: